| Research Article | ||
Open Vet J. 2023; 13(5): 558-568 Open Veterinary Journal, (2023), Vol. 13(5): 558–568 Original Research Characterization of the camel pox virus strain used in producing camel pox virus vaccineKydyrbay Maikhin1, Maxat Berdikulov1, Abdikalyk Abishov2, Yerlan Pazylov1, Gulzhan Mussayeva1, Slukyz Zhussambayeva1 Gulmira Janabekova3, Ainash Shaimbetova1, Yessengali Ussenbekov3 and Nazym Syrym4*1National Reference Center for Veterinary, Almaty, Kazakhstan 2Virology Laboratory, LLP «SPE DiaVak-ABN», Almaty, Kazakhstan 3Kazakh National Agrarian Research University, Almaty, Kazakhstan 4Laboratory of Microbiology, Research Institute for Biological Safety Problems, Almaty, Kazakhstan *Corresponding Author: Nazym Syrym. Laboratory of Microbiology, Research Institute for Biological Safety Problems, Almaty, Kazakhstan. Email: nazym_syrym31 [at] rambler.ru. Submitted: 11/12/2022 Accepted: 10/04/2023 Published: 09/05/2023 © 2023 Open Veterinary Journal
AbstractBackground: The camel pox virus (CMLV) is a widespread infectious viral disease of camels. It is necessary to conduct research on new strains for the development of vaccines. Aim: The research aims to characterize a novel strain isolated from the CMLV used to produce a CMLV vaccine. Methods: The objects of the study were the “M-0001” strain isolated from a sample of animals infected with the CMLV during the epidemic. The cultural and reproductive properties of the virus isolate were studied using primary cell lines from primary trypsinized lamb kidney and testicular cell cultures (LK and LT). Other samples included kidney cell lines from transplanted sheep as well as a kidney cell line from transplanted cattle, Vero (transplanted green monkey kidney cell line), and calf trachea. The strain was polymerase chain reaction (PCR)-tested and sequenced for characterization purposes. Results: The PCR results show that the study sample is species specific and corresponds to the CMLV by the size of the cumulative amplifications, which is 241 bp. Given the maximum percentage of a sequence match analyzed by the BLAST algorithm based on the international database and the results of phylogenetic analysis, the M0001 sample was determined to belong to the CMLV (gene bank inventory number KP768318.1). Conclusion: The sample “M0001” is located on the same branch with a representative from CMLV. Among the cell cultures tested, the LK and LT cell lines were the most sensitive to the isolated CMLV isolate. Reproducing the virus in these cell cultures remains stable even after 15 consecutive passes. The cytopathic effect of the virus was less pronounced and low in transplanted cell lines, and the cytopathic effect was no longer apparent in the third passage. A genome alignment of the virus has identified potentially conserved sites, and analysis of loci in different virus types revealed one maximally conserved locus. An epizootic strain of the camelina virus “M-0001” candidate to produce vaccines for the camels was obtained. An experimental vaccine sample based on an isolated and charred camellia virus will be created in the future. Keywords: Camel, Camel pox virus, Isolate, Primary and transplanted cell culture lines, Virus titer. IntroductionTaxonomically, Camel pox virus (CMLV) belongs to the species CMLV, genus Orthopoxvirus (OPV), a member of the large family Poxviridae, which includes viruses of humans, animals, birds, and insects. The CMLV is a rather large virion with an average size of 220 × 280 nm. As the disease only spreads to camels, unlike the CMLV, the pathogen has not been sufficiently researched. However, there is evidence that the virus is relatively resistant outside the body. As a result, it dies instantaneously after boiling and at 60°C for 15 minutes. Decomposing pathological material is rapidly inactivated, and sunlight or artificial ultraviolet irradiation destroys the pathogen within 2–3 hours (Ayelet et al., 2013; Bayisa, 2020; Barykin et al., 2021; Ussenbekov et al., 2022). CMLV is highly contagious in nature and causes serious health consequences, up to and including camel mortality and economic damage to camel owners. It manifests in either a localized mild or generalized/severe form. CMLV was first described in a region of India in the late 1900s. During the first third of the last century, this camel disease was investigated and a specific pathogen was identified. However, this disease has yet to be fully clarified. Note that CMLV is non-pathogenic for most animals, but experimental infection of rabbits and laboratory mice may occur. On this basis, a biological test is performed on camelids to establish the exact cause agent (Bayisa, 2020; OIE-WAHIS, 2020). CMLV is a widespread infectious viral disease of camels. This disease is distinctive for members of the camel family living in the Old World and those living in the New World (OIE-WAHIS, 2020). Camel pox is widespread and extends from Africa through the Middle East and Asia. The International Epizootic Bureau has found that in 2017–2021, camel pox foci were detected in 12 countries of the world. When analyzing cases over the past 5 years, it was found that in three countries (Israel, Iraq, and Eritrea), the outbreak of camel pox had sporadic cases, and the remaining nine countries (Iran, Kazakhstan, Libya, Oman, Palestine, Saudi Arabia, Somalia, Tunisia, and Ethiopia) were endemic for this disease. The CMLV causes significant economic damage in countries where camels constitute an important part of the farm animal population. CMLV, in its neglected form, may prove fatal. On the other hand, the CMLV has not been reported in Australia’s animal population (World Organization for Animal Health, 2018). Camel pox has been reported in all other countries where camel farming takes place. Symptoms of camel pox include the development of fever accompanied by lesions in the form of rash both on the skin and mucous membranes in the respiratory organs and oral cavity of animals (Marcacci et al., 2020). Depending on the severity of the disease, clinical signs of the CMLV include manifestations of different levels of systemic infection, ranging from mild to severe (Saud et al., 2021). Most commonly, the disease is cured in young and pregnant animals. The virus’s incubation period varies from 9 to 13 days, but in some cases, it may be up to 3 and 15 days. The severity of clinical symptoms may be associated with different virus strains. Furthermore, the immune status of a particular camel specimen may be significant (Aregawi et al., 2018). Besides the rash and fever, symptoms that can be detected during examination include enlarged lymph nodes. A rash can occur the day after infection or up to 3 days. The rash usually shows up after the fever. As the rash develops, erythema forms, and the papules and bladders merge into pustules. The bursting of pustules is followed by the formation of scabs. Disease localization also has its patterns. First, there’s a rash on the head, the nostrils, and the ears. The tumor may spread to the whole head for a severe form of the disease. Later, from the head, the lesions will spread to the perineum, cervical area, genital area, and breast glands (Diaz, 2021). In the worst cases, the entire body of the animal may be affected. Depending on different methods and different disease development levels, treatment can take 4–6 weeks. The transmission of the pathogen is relatively simple, generally by contact between sick and healthy animals whose immunity is not strong enough. In addition, the infection may also happen through indirect transmission through the environment. Insect transmission is also feasible, particularly after precipitation (Bulatov et al., 2010). The virus is transmitted either because of direct contact between infected and susceptible animals or through an infected environment. The infection usually enters the body through the respiratory tract or damaged areas of skin. The virus is most often transmitted through contact between animals. This is associated with frequent localization of the affected skin on the head. The virus is secreted into milk and saliva, as well as into eye and nasopharynx secretions. Dried scabs that fall off from smallpox-affected skin can contain a live virus for at least 4 months, infecting the environment. However, transmission by blood-sucking insects has not been proven yet. The incidence rate of camel pox is variable and depends on whether the virus circulates in a herd. Serological studies conducted in various countries have revealed a high prevalence of antibodies to the CMLV (Wernery and Kaaden, 2002). Morbidity rates are higher in males than in females, and mortality rates are higher in young than in adult camels (Kritz, 1982). Adult animal mortality rates range from 5% to 28%, while for young animals, it is from 25% to 100% (Mayer and Czerny, 1990). This may be due to the fact that the immunity of adult animals is at a higher level. It should be noted that the CMLV is observed in camels, while other animals remain uninfected. In 2009, there was only one human infection with the CMLV in India. At the same time, the threat to humans posed by this virus is very limited (World Organization for Animal Health, 2018). Thus, the CMLV is not virulent to humans. The incidence of the disease in camels is very variable and depends on the time the virus moves through the herd. Incidence is known to be sex-dependent and significantly higher in males, and mortality rates are higher in young camels than in adults (Mohammadpour et al., 2020). The last major camelina epizootic recorded goes back to the middle of the last century in Turkmenistan (Duraffour et al., 2011; Balamurugan et al., 2013). Despite improving the epizootic situation of CMLV in the Republic of Kazakhstan, the problem of improving livestock health has not been definitively solved. There is a need to clarify the conditions for long-term well-being and the causes of new cases of illness on healthy farms. A modern feature is the restructuring of livestock, the establishment of farms and rented farms, and the increase of livestock on private farms of citizens. The risk of introducing pathogens of the CMLV into wholesome herds has increased. The diagnosis of the CMLV may be based on clinical evidence in animals with camel pox. This includes increased saliva secretion, lacrimal secretions, and a discharge of nasopharynx pus. Severe illness includes diarrhea and anorexia. With pregnancy, a miscarriage may occur. If lethal, it can be caused by secondary infection or septicemia (sepsis without purulent metastases). Once clinical signs of the disease have emerged, tissue samples (skin or organ biopsies) are most helpful in identifying the infectious agent. Multiple investigative methods should be used to diagnose, as there are many diagnostic approaches. These include transmission electron microscopy (TEM), virus isolation by cell culture, standard polymerase chain reaction (PCR) analysis, immunohistochemistry, and neutralizing antibody demonstration. However, the identity of the pathogen as CMLV requires confirmation by TEM, PCR, and/or sequencing. TEM and restrictive enzyme analysis can be used to differentiate camel pox from other infections caused by OPV and PPV [plum pox (shark) virus]. TEM can differentiate between OPV, which is brick-shaped, and PPV, which is egg-shaped. The CMLV antigen in infected scabs and pox can be identified by immunohistochemistry. Isolation of the virus using embryonic eggs and various cell lines and primary cell cultures may be used in isolation studies. However, isolation alone cannot be the gold standard for diagnosis; serology or PCR monitoring is required (Balamurugan et al., 2013; Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 2022; Zhivotnovodstvo, 2022). Special treatment is not available. However, some antiviral drugs, such as horseradish peroxidase conjugate from rabbit serum antibodies to G-immunoglobulin of the camel, are effective in suppressing the virus in animal models (MacLachlan and Dubovi, 2011; Kandeel and Al-Mubarak, 2022). Vaccination is used for prophylactic purposes: live and inactivated vaccines have been developed worldwide (Kandeel and Al-Mubarak, 2022). An effective way to control CMLV is vaccination. Various inactivated and live attenuated CMLV vaccines are available in different countries. A live attenuated Ducapox vaccine is produced in South Africa, an inactivated T8 vaccine was developed in Morocco in 1992, and a live attenuated Jouf-78 vaccine is produced in Saudi Arabia (Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 2022). They contain the following strains: CML/Db-92, CML/Nw-wt, CML/Tm-wt, CML/Sh30-wt, CML/Sh41-wt, CMLV/43-Weight that were isolated in different years from sick camels in Sudan in different cell cultures; CMLV-Jouf (AlJouf CMLV strain isolated from sick camels in Saudi Arabia and passaged in camel kidney cell culture); Ducapox [Dubai attenuated CMLV vaccine obtained by passaging in Vero cells at the Central Veterinary Research Laboratory, Dubai, using a UAE isolate (strain CaPV298-2); Jouf-78, VD47/25, Ducapox 298/89; CMLV-1 (DNA isolated from purified CMLV-Tehran or CP-1); 6] CMLV-14 (DNA isolated from purified CMLV CP-14 isolated from camels in the United Arab Emirates) (Kandeel and Al-Mubarak, 2022). The main sources of camel pox are diseased camels or camels that have been diseased. A vaccine against the CMLV of the “M-2020” strain was developed by the CSR “Research Institute of Biological Safety Problems” (Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 2022). The significance of the studyCMLV-based vaccines have shown comparatively high efficacy (World Organization for Animal Health, 2018). Attenuated vaccines are more effective, their induced immunity is more protracted than inactivated vaccines. Attenuated vaccines are vaccines obtained by weakening a pathogenic virus or bacteria in the laboratory. At the same time, their widespread use in virus-free areas is associated with elevated risks of spreading mesogenic virus strains in a healthy population. Medium virulence strains are mesogenic that can be accompanied by a decrease in camel productivity (reduction in milk production and the rate of weight gain of animals) (Strognova and Trukhonenko, 2013). Thus, the absence of a cost-effective vaccine against the CMLV in the Republic of Kazakhstan and many camel-breeding countries is a major obstacle to the fight against disease outbreaks. This study characterizes a newly isolated strain of CMLV used for vaccine production. There are 31 million cattle in Kazakhstan and 214 thousand of them are camels. The CMLV in Kazakhstan and its epidemiology have not been studied in Kazakhstan. Therefore, Kazakhstan is a convenient model territory for studying the CMLV and searching for new strains. The aim of the studyThe study aimed to examine the characteristics of the novel isolated CMLV strain. The study’s objectives included: a) isolating the strain; b) performing cytotoxic tests of this strain on different cell cultures; c) establishing the characteristics of the strain by sequencing to enter it into the database. This work’s results will help optimize the anti-epizootic measures to prevent the emergence and spread of this disease in the Republic of Kazakhstan camels. Materials and MethodsResearch regionThe study was conducted in 2021–2022 in the Almaty region of Kazakhstan. CMLV cultivationCultural and reproductive properties of the CMLV isolate were analyzed on primary cell culture lines of primary trypsinized lamb kidney and testicular cell culture (LK and LT) and finite cell lines of sheep kidney (SK), a finite cell line of the bovine kidney (BK), Vero (finite cell line of green monkey kidney), and calf trachea (CT). LT; LK; goats’ testicles; and goats’ kidneys (GK). A 20% suspension prepared from organic material (scratches of the camel pox lesions on the skin) isolated from 27 CMLV-infected animals of the Mangistau region was used for cultivation and adaptation. The sensitivity of the primary and transplanted cell lines to the isolate was verified in three consecutive runs. The nutritious medium was drained from selected cell cultures. Some of the cultures (at least 2) were left as controls, to which 1 ml of the maintenance medium was added. The remaining four cell monolayer matrices were each injected with 1 ml of virus-containing material. All cultures in the flat (control and experimental) were placed in an incubator for 1 hour at 37°C. The inoculum was drained, and the monolayer was washed with the Hanks solution several times. Afterward, the inoculum was replaced with a fresh medium and grown at the same temperature for 10 to 12 days. During incubation, the mattress environment was changed every 3 days. Monolayer microscopy was performed daily during the indicated period to detect the virus’s CPE (Fig. 1). From the selected biological material, a 20% tissue suspension was applied to a sterile physiological solution, frozen and thawed three times, and centrifuged at 3,500 revolutions per minute for 30 minutes. The supernatant was transferred to sterile specimens and incubated at room temperature for 30 minutes after adding antibiotics. The supernatant was subsequently examined for sterility. Afterward, the isolated isolate was subjected to a virological examination to study its cultural properties. The growing medium was removed from the vessels through cell culture to infect cell cultures with the virus. The viral suspension was introduced at an appropriate dose, and the single-layer surface was slightly swayed and incubated at 37°C for 1 hour. The inoculum was drained, the monolayer of the infected cell culture was filled with a supporting medium, and the cultivation continued at 37°C. After infection, the viral mass was collected after the maximal development of CPE in the single layer. To that end, the cell culture in the mattresses was frozen at minus 200°C, and thawed at room temperature while shaking intensively. The contained viral suspension was placed in sterile containers of 150 to 200 ml and stored at −20°C in the frozen state until it was used for work. The virus titer in the culture fluid containing the virus was determined through titration in a test tube cell culture. For this purpose, 10-fold dilutions (10−1–10−7) were prepared from the studied suspension on a support medium, and four test tube cell cultures were infected with each suspension dilution in a dose of 1 cm3. The cytopathogenic activity of the virus was assessed based on the timing of onset, the intensity of CPE development, and the accumulated virus titer. Primary and translucent cell lines grown in test tubes and matrices were used for growing the CMLV isolate. Cell cultures were grown in Igla-MEM, a DMEM medium containing 10% bovine serum, and preserved in the same medium containing 2% bovine serum. Cell and virus cultures were cultured at 37°C stationary. The susceptibility of cell cultures to the CMLV isolate in each assay was assessed at the time of CPE development. The intensity of its development and the virus titer at the end of cultivation. At the time of maximum CMLV development (80% or more of the cell monolayer area) or in the case of no manifestation and weak effect, they were frozen at minus 40°C on 10th–12th days after infection for the subsequent successive passages. Then they were thawed at room temperature, combined in separate matrices, and the resulting suspension was used to infect a fresh similar cell culture after checking for sterility (MPB and MPA, Fig. 1). The methods of infection, cultivation, virus collection, sensitivity evaluation, and cell culture adaptation in the second and subsequent passages are similar to those for the first passage (Strognova and Trukhonenko, 2013; Online.zakon, 2022). Determination of CMLV biological activity titerCell cultures: (Vero, GK, CT, BK, LK, and LT,) were plated on 96-well plates 24 hours before infection. The CMLV supernatants were diluted 10-fold by serial dilution (101 to 107), and 200 μl of each dilution was seeded in six wells. A cell culture medium without the virus was used as a control. Infected plates with cell cultures were incubated at 37°C for 5–7 days, depending on the duration of CDC manifestation; the virus titer was calculated in lg TCD 50/сm3. The virus titer was calculated according to the method of Reed and Mench. The nutrient medium in the plates was changed every 2–3 days depending on the pH of the medium (VMEDE, 2015; Farmf, 2022).
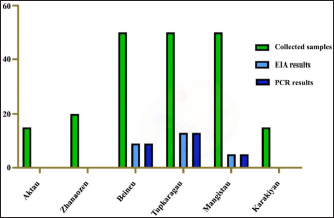
Fig. 1. Cell culture adaptation. (a) Cell culture flattening and matting. (b) Counting cell cultures for sieving in a Goryaev’s chamber. (c) Grown cell cultures in flats. Information and software utilities “BLAST” and “Vector NTI 9.1.0” were used to study molecular genetic characteristics. Plasmid DNA with a marker insert was used for PCR amplification. According to the manufacturer’s recommendations, viral DNA was isolated using the PureLink Microbiome DNA kit (Invitrogen) (Biochemmack, 2022). DNA fragmentation will be performed to a size of about 400–450 bp using a DNA Fragmentation kit (NEB, USA). According to the attached protocol, libraries for parallel mass sequencing were prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina (NEB, USA). The quality of the prepared libraries will be tested using a Bioanalyzer 2,100 (Agilent Technologies, Germany) (New England BioLabs, 2021). Sequencing will be performed using the MiSeq Reagent v.3 kit (Illumina, USA) on a new generation MiSeq sequencer (Illumina, USA). Information and software utilities “BLAST” and “Vector NTI 9.1.0” were used to study the molecular genetic characteristics of isolated isolates. Plasmid DNA with a marker insert was used for PCR amplification. PCR products were purified from unbound primers by enzymatic method using Exonuclease I (Fermentas) and Shrimp Alkaline Phosphatase (Fermentas). Bioinformatic analysis of the sequences obtained as a result of sequencing will be performed using Geneious 11.0 computer software (Biomatters, New Zealand) (Genious, n.d.). The sequencing reaction was performed using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer’s instructions, followed by fragment separation on a 3,730 × l DNA analyzer (Applied Biosystems) The nucleotide sequences were analyzed and combined into a common sequence using the SeqMan software (DNAStar). After that, end fragments (nucleotide sequences of primers, and fragments with a low-quality score) were removed. The resulting sequences were identified in GeneBank using the BLAST algorithm. The amplified target DNA fragments were analyzed by separating DNA fragments in 1.5% agarose gel in the presence of an intercalating agent, ethidium bromide, which was used for further DNA visualization. Electrophoresis was performed in a PowerPac horizontal electrophoresis chamber using a BioRad Electrophoretic bath current source, and 1-TAE buffer was used as an electrode buffer. Alignment and phylogenetic analysis of sequenced genes with nucleotide sequences from GenBank will be performed using the MEGA 6.0 computer program using the maximum likelihood method based on 500 samples, GTR model. Taking into account the databases indicating the availability in international nucleotide sequence banks GeneBank (http://www.ncbi.nlm.nih.gov/), Ribosomal Database Project (http://rdp.cme.msu.edu/html/), we additionally performed the construction of phylogenetic trees with nucleotide sequences of ATI genes of reference strains of these species. The nucleotide sequences of the ATI genes phylogenetically, and most related microorganisms were included in the analysis. The Mega 11 software was used to construct phylogenetic trees. The Muscle algorithm was used to align the nucleotide sequences; the ancient trees were constructed using the Neighbor - JoiningNJ method. Ethical approvalThe authors declare that the work is written with due consideration of ethical standards. The study was conducted in accordance with the ethical principles approved by the Ethics Committee of Research Institute for Biological Safety Problems (Protocol No. 6 of 13.08.2022). ResultsDuring an outbreak of the CMLV in the Mangistau region in 2020, employees of the “National Reference Center for Veterinary Medicine” collected paired blood samples for EIA (enzyme immunoassay) and PCR, and samples were taken from the CMLV lesions of skin from sick camels for PCR. The results of the studies are presented in Table 1. Tabel 1. PCR and EIA results for CMLV.
Fig. 2. Mangistau region. As follows from Table 1 and Figure 2, all 27 samples from the Mangistau region (Beineu, Tupkaragan, Mangistau districts) tested positive by EIA and PCR methods. Table 2 presents the determination of the CMLV susceptibility for various primary and finite cell cultures. As seen from Table 2 to Figure 3, in primary infections, the CPE of the virus in LK and LT cell cultures ranged between 1.50 and 2.25 lg 50 cm3 TCD from the first to the third passages. In finite cultures of GK, CT, Vero, and BK cells, similar changes were noted later by 1–2 days, they developed relatively slowly. On the 7th and 8th days, the cell monolayer area with CPE reached only 30%–40%. The intensity of CPE of the pathogen in the second passage in the above finite cell lines was less noticeable and weak, and in the third passage, CPE was no longer evident. As seen in Figure 3, the cultures of GK, CT, Vero, and BK cells showed similar changes later by 1–2 days, they developed relatively slowly, and on day 7–8, the cell monolayer area with CPE reached 30%–40%. The intensity of CPE of the pathogen in the second passage in the above finite cell lines was less noticeable and weak, and in the third passage, CPE was no longer evident. The CMLV in 15 consecutive passages reproduced and accumulated in a titer of 6.00–6.25 lg TCD 50 cm3 in both LK and LT cell cultures. The cytopathogenic effect of the virus in the monolayer of infected cells was clear 72–96 hours after infection and covered more than 85% of its area between 110 and 144 hours. Thus, the results of the experiments show that the CMLV among the tested primary and transplanted cell cultures was the most sensitive of the LT and LK cell lines (primary trypsinized culture). Virus reproduction in these cell cultures remains stable even after 15 consecutive passages. PCR and DNA electrophoresis. DNA extraction from the virus-containing suspension was performed using the Blood and Tissue Kit (Qiagen) according to the manufacturer’s procedures. The results are shown in Figure (4). Table 2. Determination of the sensitivity of the CMLV to various primary and finite cell cultures.
As shown in Figure (4A), 887 bp were accumulated with the generic primers to the ATIP locus of OPVs, which confirms the detection of CMLV. According to the size of the accumulated amplification of 241 bp in Figure (4B), the sample under study is species-specific and consistent with the CMLV. The nucleotide sequences of the CMLV viral genome were analyzed. Alignment of the virus genome with potential conserved sites. Comparing these loci in different virus types revealed one maximally conserved locus. Subsequent BLAST analysis demonstrated the high genome specificity of the CMLV; primers and a probe were selected for this locus. Furthermore, oligonucleotide primers were chosen for the three genes that are part of the CMLVgenome, the least homologous of specific oligonucleotides. The primers and probe served as an internal amplification check. Positive control with the nucleotide sequence of the CMLVgenome marker area was designed to control the progression of amplification. A high degree of polymorphism was found in the virus genome compared to the PCR probe. Changes to the PCR probe allow us to eliminate the effect of this variability in the number of viruses. The nuclear sequences of the primers, probes, and positive controls are presented in the tables.
Fig. 3. Virus titre by passages.
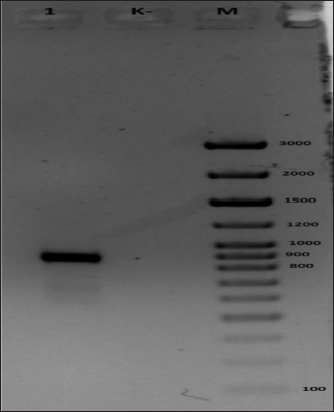
Fig. 4. Results of electrophoretic analysis in 1% agarose gel of amplification products using primers to CMLV DNA. (A) Generic primers to the ATIP locus of OPVs and (B) Species-specific primers to the C18LCMLV locus. The following samples were analyzed: 1, test DNA sample; 2, negative control; 3, control DNA sample containing CMLV DNA confirmed by sequencing; M, 1kb-plus DNA marker (Thermo Fisher Sci). DNA sample derived from the isolate was identified by determining the direct nucleotide sequence of the ATI gene fragment. Afterward, the nucleotide identity was determined with sequences deposited in the international GenBank database. The PCR reaction was performed with universal primers: - F 5′ - AATACAAGGAGGATCT-3 and OIE_R - 5′ CTTAACTTTTTTTTTTCTC-3 in a total volume of 30 μl. The PCR mixture contained 15 ng of DNA, 1 unit of Taq DNA Polymerase (Fermentas), 0.2 mM of each dNTP, 10×*KCl buffer (Fermentas), 2.5 mM MgCl2, 10 pmol of each primer. The PCR amplification program included a long denaturation at 95°C for 6 minutes; 35 cycles: at 94°C for 30 seconds, at 55°C for 30 seconds, at 72°C for 1 minute; a final elongation of 9 minutes at 72°C; the PCR program was performed using a SimpliAmp Thermal Cycler amplifier (Applied Biosystems). A specific fragment was amplified in the test sample. No PCR product was observed in the negative control, indicating the absence of contamination (Fig. 5). The results of sequencing are shown in Table 3. As illustrated in Figure (6), the sample “M0001” is on the same branch with a representative of CMLV. Considering the maximum percentage correspondence of the analyzed sequence in the international BLAST database and the results of phylogenetic analysis, the sample “M0001” was determined as belonging to the CMLV. For example, a CMLV strain, “M0001,” a candidate strain for producing a CMLVvaccine, was cultivated. The CMLV strain used to accumulate the pathogen biomass was obtained by reproducing it in a monolayer culture of LK and LT cells at a titer of 6.00–6.25 lg TCD 50 cm3, respectively, maintained under laboratory conditions.
Fig. 5. PCR electropherogram of amplification products. 1. Test sample; (M) Step 100 long molecular weight marker, (C-) negative control sample. DiscussionThe CMLVis registered in virtually all countries where camel farming is practiced. During epizootic outbreaks of the CMLV, adult mortality can reach 10%, camel abortions up to 25%–27%, and litter loss up to 85% and higher. The CMLV is diagnosed based on epizootic data, characteristic symptoms of the disease, positive results of microscopy (processing of smears and fresh papules), electron microscopy, as well as the results of laboratory tests: PCR, restriction analysis, EIA (ELISA) (Diaz, 2021). A wide range of serological tests, including neutralization, precipitation, hemagglutination, hemagglutination inhibition, complement binding, fluorescent antibodies, and ELISA, are used to detect postvaccination antibodies against the CMLV and antibodies after natural disease transmission. According to Kazakhstan’s veterinary reporting data, the CMLV epizootics were observed periodically in Mangistau and Atyrau regions in 1930, 1942–43, 1965–1969, 1996, and 2019–2020 (Bulatov et al., 2010). The evaluation of statistical data on the incidence of camel dander in the above regions of Kazakhstan has established the cyclicality of epizootics, which is about 10 to 20 years. In the period between epizootics, the infection in camels in stationary zones free of the CMLV occurs in the form of enzootics and sporadic cases that occur more or less regularly every 3–6 years, mainly among animals aged 2–4 years (Bulatov et al., 2010; Zhugunissov et al., 2021). Until recently, this disease was considered of little importance, but in this decade, it is considered an emerging public health problem due to the increasing number of reported cases and outbreaks in camels (OIE-WAHIS, 2020). It is consistent with data from other authors. For instance, both live and attenuated vaccines were found to be similar in terms of efficacy (Dahiya et al., 2016). It has also been shown in this research, using the cytotoxicity of the virus to different cell cultures as an example. Moreover, the pathogenesis of the virus is intended to suppress the host’s immune system, which is the cause of elevated lethality (Dahiya et al., 2016). Antibodies were discovered in an in-depth study in 19% of camels and 81% of herds examined (Aregawi et al., 2018). This study’s authors attribute the virus’s highly contagious nature to the extensive mixing of camels in the herd. Examples of prevention include improved housing conditions and vaccination (Aregawi et al., 2018). The vaccine we are developing can also contribute to infection control. Novel isolated subtypes in the CMLVdatabase, similar to the strain we studied, have been found in the Israeli camel population (Erster et al., 2018). At the same time, some isolated te CMLV strains were similar to cowpox strains (Khalafalla et al., 2020). Table 3. Results of identification by nucleotide sequence analysis of the ATI gene.
Fig. 6. Phylogenetic tree based on analysis of the AT fragment from sample M0001. A consequence of this may be the cowpox vaccine’s efficacy in the CMLVcases (Haagmans et al., 2016). Therefore, the proximity factor for individual viruses must also be considered (MacNeill, 2022). Some authors, noting the need to develop new vaccines, also point to their stable immune response to coronaviruses, which can be the basis for developing vaccines already against coronaviruses (Kandeel and Al-Mubarak, 2022). Despite single cases of the human CMLV, new cases of infection were already reported in 2014 in Sudan (Khalafalla and Abdelazim, 2017; ICTV, 2019). The infectious titer obtained on the skin of vaccinated animals was at least 1.5 log of semi-infecting doses, which is less than the titer obtained on the skin of control animals. In addition, vaccinated animals did not have any common symptoms of the disease (Haagmans et al., 2016). The effectiveness of the inactivated vaccine was also confirmed by the post-vaccination production of antibodies evaluated by methods of solid-phase ELISA and virus neutralization. There are no vaccines that allow the implementation of the DIVA strategy (detecting infection in vaccinated animals). The development of long-term immunity provided by the inactivated vaccine was also confirmed by infecting vaccinated animals 1 year after the initial vaccination. The duration of immunity provided by the inactivated vaccine is at least 1 year after the administration of two doses of the vaccine. This was confirmed by a test conducted on young untreated single-humped camels (Aregawi et al., 2018). After repeated vaccination, the duration of immunity may be longer in adult single-humped camels (Dahiya et al., 2016). There are no reports of unsuccessful field vaccinations of animals that do not receive regular vaccination. In this regard, special attention should be paid to the CMLVoutbreaks in camels. Molecular biological characterization studies, enhanced diagnostic methods, and preventive, and control measures are of paramount importance in reducing the circulation of CMLV in camels. Effective preventative and control measures can be implemented using appropriate diagnostic and prophylactic tools to contain the CMLVspread, as described for most zoonoses (World Organization for Animal Health, 2018). ConclusionThe CMLV “M-0001” has been isolated from lesions of a diseased camel pox. The sample “M0001” is located on the same branch with a representative from CMLV. Thus, the results of the experiments show that the CMLVwas the most sensitive of the LK and LT cell lines (primary trypsinized culture) among the tested primary and finite cell cultures. Reproducing the virus in these cell cultures remains stable even after 15 consecutive passes. A genome alignment of the virus has identified potentially conserved sites, and analysis of loci in different virus types revealed one maximally conserved locus. The subsequent BLAST analysis determined that it was highly specific to the CMLVgenome; primers and a probe were selected for this location. In the future, an experimental sample vaccine will be created from isolated and characterized the CMLV. New, highly sensitive, and specific methods involving genomics and conventional methods will be useful in identifying new and emerging viruses. In the future, it is necessary to conduct the experimental implementation of the resulting vaccine on a larger scale to test its effectiveness. It is also necessary to constantly monitor new camel pox strains. The research limitations of the study imply that the developed vaccine is still at the experimental stage and its implementation requires tests. The data obtained for Kazakhstan can also be applied to the livestock of camels in other countries where camel breeding is intensively developed. The results of genetic identity can be used as a molecular-biological characterization of the isolated strain of the CMLV. Conflict of interestThe authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. FundingThe authors received no financial support for the research, authorship, and/or publication of this article. ReferencesAregawi, W.G., Agga, G.E., Gishe, J. and Abdi, R.D. 2018. Seroprevalence and participatory epidemiology of camel pox virus in Afar region of Ethiopia. Prev. Vet. Med. 161, 25–32. Ayelet, G., Jenberie, S., Belay, A., Mohammed, A., Mola, B., Gizaw, Y., Muhie, Y., Gelaye, E., Asmare, K. and Skjerve, E. 2013. The first isolation and molecular characterization of Camel pox virus in Ethiopia. Antivir. Res. 98(3), 417–422. Balamurugan, V., Venkatesan, G., Bhanuprakash, V. and Singh, R.K. 2013. Camel pox virus, an emerging orthopox viral disease. Indian. J. Virol. 24(3), 295–305. Barykin, S.E., Kapustina, I.V., Korchagina, E.V., Sergeev, S.M., Yadykin, V.K., Abdimomynova, A. and Stepanova, D. 2021. Digital logistics platforms in the BRICS countries: comparative analysis and development prospects. Sustainability 13(20), 11228. Bayisa, D.A. 2020. Review on camel pox virus: epidemiology, public health and diagnosis. ARC. J. Anim. Vet. Sci. 5(4), 22–33. Biochemmack. 2022. PureLink genomic DNA mini kit. Available via https://www.biochemmack.ru/catalog/element/14314/29494/ (Accessed 10 September 2022). Bulatov, E.A., Mamadaliev, S.M., Mambetaliev, M. and Bitov, N.T. 2010. Circulation of camel pox virus in Manghystayskaya Region, Republic of Kazakhstan, in the latent form. Actual. Issues. Vet. Biol. 3(7), 10–13. Dahiya, S.S., Kumar, S., Mehta, S.C., Narnaware, S.D., Singh, R. and Tuteja, F.C. 2016. Camel pox virus: a brief review on its epidemiology, current status and challenges. Acta. Trop. 158, 32–38. Diaz, J.H. 2021. The disease ecology, epidemiology, clinical manifestations, management, prevention, and control of increasing human infections with animal orthopoxviruses. Wilderness. Environ. Med. 32(4), 528–536. Duraffour, S., Meyer, H., Andrei, G. and Snoeck, R. 2011. Camel pox virus. Antivir. Res. 92(2), 167–186. Erster, O., Melamed, S., Paran, N., Weiss, S., Khinich, Y., Gelman, B., Solomony, A. and Laskar-Levy, O. 2018. First diagnosed case of camel pox virus in Israel. Viruses 10(2), 78. Farmf. 2022. Cultivation of viruses. Methods. Available via https://farmf.ru/lekcii/kultivirovanie-virusov-metody/ (Accessed 10 September 2022). Genious, n.d. Discover the world’s leading bioinformatics software platform. Available via https://www.geneious.com/ (Accessed 10 September 2022). Haagmans, B.L., van den Brand, J.M., Raj, V.S., Volz, A., Wohlsein, P., Smits, S.L., Schipper, D., Bestebroer, T.M., Okba, N., Fux, R., Bensaid, A., Solanes Foz, D., Kuiken, T., Baumgärtner, W., Segalés, J., Sutter, G. and Osterhaus, A.D. 2016. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science 351(6268), 77–81. ICTV. 2019. ICTV International committee on taxonomy of viruses. Available via ictvonline.org (Accessed 10 September 2022). Kandeel, M. and Al-Mubarak, A. 2022. Camel viral diseases: current diagnostic, therapeutic, and preventive strategies. Front. Vet. Sci. 9, 915475. Khalafalla, A.I. and Abdelazim, F. 2017. Human and dromedary camel infection with camel pox virus in Eastern Sudan. Vector. Borne. Zoonotic. Dis. 17, 281–284. Khalafalla, A.I., Al Hosani, M.A., Ishag, H. and Al Muhairi, S.S. 2020. More cell culture passaged Camel pox virus sequences found resembling those of vaccinia virus. Open. Vet. J. 10(2), 144–156. Kritz, B. 1982. A study of camel pox in Somalia. J. Comp. Pathol. 92(1), 1–8. MacLachlan, N.J. and Dubovi, E.J. 2011. Poxviridae: camel pox virus. Fenner’s veterinary virology, 4th ed. London, UK: Academic Press, pp: 151–165. MacNeill, A.L. 2022. Comparative pathology of zoonotic orthopoxviruses. Pathogens (Basel, Switzerland) 11(8), 892. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 2022. Available via https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/A_summry.htm (Accessed 10 September 2022). Marcacci, M., Khalafalla, A.I., Al Hammadi, Z.M., Monaco, F., Cammà, C., Yusof, M.F., Al Yammahi, S.M., Mangone, I., Valleriani, F., Alhosani, M.A., Decaro, N., Lorusso, A., Almuhairi, S.S. and Savini, G. 2020. Genome sequencing of a camel pox virus vaccine reveals close similarity to modified vaccinia virus Ankara (MVA). Viruses 12(8), 786. Mayer, A. and Czerny, C.P. 1990. Chapter 4, Camel pox virus. In Virus infections of vertebrates. Virus infections of ruminants. Eds., Dinter, Z. and Morein, B. Amsterdam, The Netherlands: Elsevier Science Publisher, pp: 19–22. Mohammadpour, R., Champour, M., Tuteja, F. and Mostafavi, E. 2020. Zoonotic implications of camel diseases in Iran. Vet. Med. Sci. 6(3), 359–381. New England BioLabs. 2021. NEBNext® Ultra™ DNA Library Prep Kit for Illumina®. Available via https://international.neb.com/products/e7370-nebnext-ultra-dna-library-prep-kit-for-illumina#Product%20Information (Accessed 10 September 2022). OIE-WAHIS. 2020. Disease situation: Camel pox virus. Available via https://wahis.oie.int/#/dashboards/country-or-disease-dashboard (Accessed 10 September 2022). Online.zakon. 2022. Cultivation of viruses. Available via https://online.zakon.kz (Accessed 10 September 2022). Saud, Z., Hitchings, M.D. and Butt, T.M. 2021. Nanopore sequencing and de novo assembly of a misidentified camel pox virus vaccine reveals putative epigenetic modifications and alternate protein signal peptides. Sci. Rep. 11, 17758. Strognova, I.Y. and Trukhonenko, A.A. 2013. The use of cell culture in virology: methodological quidelines. Krasnoyarsk, Russia: Krasnoyarsk State Agrarian University. Available via http://www.kgau.ru/sveden/2017/ipbivm/mu_360501_9.pdf (Accessed 10 September 2022). Ussenbekov, Y., Bagdat, A., Bimenova, Z., Orynkhanov, K., Sobiech, P., Samardžija, M., Pareek, C.S. and Dobos, A. 2022. Identification of monomorphic and polymorphic genes associated with recessive fertility defects in Holstein cows reared in Kazakhstan. Vet. Arh. 92(1), 27–35. VMEDE. 2015. Methods for culturing viruses. Available via http://vmede.org/index.php?Topic=584.0 (Accessed 10 September 2022). Wernery, U. and Kaaden, O.R. 2002. Camel pox. In Infectious diseases in camelids, 2nd ed. Eds., Wernery, U. and Kaaden, O.R. Berlin, Germany: Blackwell Science, pp: 176–185. World Organization for Animal Health (OIE). 2018. Camel pox virus (Vol. 2. Chapter 3.9.2). Available via https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.09.02_CAMEL POX VIRUS.pdf (Accessed 10 September 2022). Zhivotnovodstvo. 2022. Camel pox virus variola camelina. Available via http://zhivotnovodstvo.net.ru/maloizvestnye-zaraznye-bolezni-zhivotnyh/1962-ospa-http://zhivotnovodstvo.net.ru/maloizvestnye-zaraznye-bolezni-zhivotnyh/1962-ospa-verblyudov-variola-camelina.html">verblyudov-variola-camelina.html (Accessed 10 September 2022). Zhugunissov, K., Kilibayev, S., Mambetaliyev, M., Zakarya, K., Kassenov, M., Abduraimov, Y., Bulatov, Y., Azanbekova, M., Absatova, Z., Abeuov, K., Nurgaziev, R., Renukaradhya, G.J. and Tabynov, K. 2021. Development and evaluation of a live attenuated egg-based camel pox virus vaccine. Front. Vet. Sci. 16(8), 721023. | ||
| How to Cite this Article |
| Pubmed Style Maikhin K, Berdikulov M, Abishov A, Pazylov Y, Mussayeva G, Zhussambayeva S, Janabekova G, Shaimbetova A, Ussenbekov Y, Syrym N. Characterization of the camel pox virus strain used in producing camel pox virus vaccine. Open Vet J. 2023; 13(5): 558-568. doi:10.5455/OVJ.2023.v13.i5.8 Web Style Maikhin K, Berdikulov M, Abishov A, Pazylov Y, Mussayeva G, Zhussambayeva S, Janabekova G, Shaimbetova A, Ussenbekov Y, Syrym N. Characterization of the camel pox virus strain used in producing camel pox virus vaccine. https://www.openveterinaryjournal.com/?mno=93653 [Access: April 27, 2024]. doi:10.5455/OVJ.2023.v13.i5.8 AMA (American Medical Association) Style Maikhin K, Berdikulov M, Abishov A, Pazylov Y, Mussayeva G, Zhussambayeva S, Janabekova G, Shaimbetova A, Ussenbekov Y, Syrym N. Characterization of the camel pox virus strain used in producing camel pox virus vaccine. Open Vet J. 2023; 13(5): 558-568. doi:10.5455/OVJ.2023.v13.i5.8 Vancouver/ICMJE Style Maikhin K, Berdikulov M, Abishov A, Pazylov Y, Mussayeva G, Zhussambayeva S, Janabekova G, Shaimbetova A, Ussenbekov Y, Syrym N. Characterization of the camel pox virus strain used in producing camel pox virus vaccine. Open Vet J. (2023), [cited April 27, 2024]; 13(5): 558-568. doi:10.5455/OVJ.2023.v13.i5.8 Harvard Style Maikhin, K., Berdikulov, . M., Abishov, . A., Pazylov, . Y., Mussayeva, . G., Zhussambayeva, . S., Janabekova, . G., Shaimbetova, . A., Ussenbekov, . Y. & Syrym, . N. (2023) Characterization of the camel pox virus strain used in producing camel pox virus vaccine. Open Vet J, 13 (5), 558-568. doi:10.5455/OVJ.2023.v13.i5.8 Turabian Style Maikhin, Kydyrbay, Maxat Berdikulov, Abdikalyk Abishov, Yerlan Pazylov, Gulzhan Mussayeva, Slukyz Zhussambayeva, Gulmira Janabekova, Ainash Shaimbetova, Yessengali Ussenbekov, and Nazym Syrym. 2023. Characterization of the camel pox virus strain used in producing camel pox virus vaccine. Open Veterinary Journal, 13 (5), 558-568. doi:10.5455/OVJ.2023.v13.i5.8 Chicago Style Maikhin, Kydyrbay, Maxat Berdikulov, Abdikalyk Abishov, Yerlan Pazylov, Gulzhan Mussayeva, Slukyz Zhussambayeva, Gulmira Janabekova, Ainash Shaimbetova, Yessengali Ussenbekov, and Nazym Syrym. "Characterization of the camel pox virus strain used in producing camel pox virus vaccine." Open Veterinary Journal 13 (2023), 558-568. doi:10.5455/OVJ.2023.v13.i5.8 MLA (The Modern Language Association) Style Maikhin, Kydyrbay, Maxat Berdikulov, Abdikalyk Abishov, Yerlan Pazylov, Gulzhan Mussayeva, Slukyz Zhussambayeva, Gulmira Janabekova, Ainash Shaimbetova, Yessengali Ussenbekov, and Nazym Syrym. "Characterization of the camel pox virus strain used in producing camel pox virus vaccine." Open Veterinary Journal 13.5 (2023), 558-568. Print. doi:10.5455/OVJ.2023.v13.i5.8 APA (American Psychological Association) Style Maikhin, K., Berdikulov, . M., Abishov, . A., Pazylov, . Y., Mussayeva, . G., Zhussambayeva, . S., Janabekova, . G., Shaimbetova, . A., Ussenbekov, . Y. & Syrym, . N. (2023) Characterization of the camel pox virus strain used in producing camel pox virus vaccine. Open Veterinary Journal, 13 (5), 558-568. doi:10.5455/OVJ.2023.v13.i5.8 |