| Research Article | ||
Open Vet. J.. 2025; 15(8): 3809-3822 Open Veterinary Journal, (2025), Vol. 15(8): 3809-3822 Research Article Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genesMohammad Allam1, Sara Gamal Abd Allah Eisa2,*, Ali Hussein Abu Almaaty3 and Mohamed Kamel Hassan41Associate Professor of Molecular Genetics, Zoology Department, Faculty of Science, Luxor University, Luxor, Egypt 2Biotechnology Researcher, Zoology Department, Faculty of Science, Port Said University, Port Fuad, Egypt 3Professor of Genetics, Zoology Department, Faculty of Science, Port Said University, Port Fuad, Egypt 4Professor of Molecular Biology, Zoology Department, Faculty of Science, Port Said University, Port Fuad, Egypt *Corresponding Author: Sara Gamal Abd Allah Eisa,. Biotechnology Rresearcher, Zoology Department, Faculty of Science, Port Said University, Port Fuad, Egypt. Email: saragamal2421 [at] yahoo.com Submitted: 03/03/2024 Revised: 29/06/2025 Accepted: 20/07/2025 Published: 31/08/2025 © 2025 Open Veterinary Journal
ABSTRACTBackground: Study of five Carangid species to estimate the degree of genetic divergence and draw phylogenetic relationships by using cytochrome c oxidase subunit I (COI), and small (12S rRNA) mitochondrial rRNA genes. Aim: This investigation was designed to evaluate genetic relationships and association analyses in the taxonomy studies of Carangid fishes using mitochondrial sequences. Methods: The present study analyzed sequence data using two genes to estimate the relationships among five species of the family Carangidae (ray-finned fish), such as Carangoides bajad (gold-spotted trevally), Carangoides malabaricus (Malabar trevally), Caranx melampygus (Bluefin trevally), Caranx sexfasciatus (Bigeye trevally), and Scomberoides lysan (doublespotted queenfish) and to assess the phylogenetic utility of these markers. Results: The classification analysis of the family Carangidae is controversial. Our study was performed to examine the phylogenetic relationships among five Carangid species using 12S rRNA and COI genes, that illustrated certain Carangidae family genera are not monophyletic that does not include all the descendants of a common ancestor (Paraphyletic) refers to a taxonomic grouping that includes a common ancestor and some, but not all, of its descendants. This means that a paraphyletic group consists of the last common ancestor and excludes certain lineages that are part of the broader group, for example, in traditional taxonomy, the class of fish is considered paraphyletic because it does not include all descendants. The data reported here may be employed in study and analysis of the phylogenetic variety and relationships among species and genera of the family Carangidae. Conclusion: Our results confirmed the thermostability and environmental adaptation of the five species of the Carangidae family due to higher A+T content. Our results also confirmed the earlier conclusions of other authors that several genera of the Carangidae family are not monophyletic which does not include all the descendants of a common ancestor (Paraphyletic) and demonstrated the usefulness of the 12S rRNA gene and the COI gene in the phylogenetic analysis of the Carangid species. Keywords: Carangid species, Mitochondrial rRNA genes, Cytochrome c oxidase subunit I (COI), Small (12S rRNA) mitochondrial rRNA genes, Phylogenetic analysis, Environmental adaptation, Non-monophyletic. IntroductionThe carangids represent an important group of exploited pelagic coastal resources of the world with diverse morphological features comprising approximately 147 species (Lin and Shao, 1999). They inhabit a range of environments in all tropical and subtropical oceans, including pelagic, benthopelagic, and reef-associated areas (Laroche et al., 1984; Honebrink, 2000). There are currently four recognized subfamilies in the Carangidae: Trachinotini, Scomberoidini, Naucratini, and Carangini (Gushiken, 1988). Marine fishes of the Carangidae family are significant both economically and environmentally (Li et al., 2020). This family, which includes 148 species belonging to 32 genera, is one of the most economically significant coastal pelagic fish species worldwide. Carangidae are highly valuable economic species that are significant to the ecology (Thu et al., 2019). Within the order Perciformes, there are multiple families. The Carangidae family is well-known for its members in order to have significance in waters that are both tropical and subtropical. The carangid species are enormous, powerful, carnivorous fish that swim in open water (Randall, 1995; Jawad and Al-Mamry, 2012). Significant changes in morphology and pigmentation occur during growth in carangids (Bohlke and Chaplin, 1993). These changes have likely led to misidentification of samples and contributed to general taxonomic bewilderment (Jaafar et al., 2012). Often, morphological identification has some difficulties, because of inter- and intraspecific variation, while molecular markers can reliably, accurately and rapidly identify species as well as variants and cryptic taxa (Holland et al., 2004; Le Roux and Wieczorek, 2009; Garcia-Morales and Elias-Gutierrez, 2013). Recent advancements in species identification have led to the development of numerous DNA-based methods, such as restriction enzyme digestion, DNA hybridization, random PCR amplification, and sequencing of DNA (Hayashi et al., 1985; Irwin et al., 1991; Cano et al., 1993; Chow et al., 1993; DeSalle et al., 1993; Esposti et al., 1993; Collura and Stewart, 1995; Brown et al., 1996; Birstein and DeSalle, 1998; Burgener and Hubner, 1998). A critical assessment of all these techniques should concentrate on their reproducibility and discriminatory capabilities. The creation of precise and trustworthy technology for the quick screening of DNA sequence variants is one of contemporary biology’s greatest triumphs (Yang et al., 2014). Because the protein-coding (COI) gene and the mitochondrial 16S rRNA gene are substantially conserved, mitochondrial DNA has been widely examined in fish phylogenetics. Numerous taxa of vertebrates and invertebrates have had their mitochondrial genes sequenced at this time (Brown, 1985; Bermingham and Lessios, 1993; Santos et al., 2003; Munasinghe et al., 2004; Vinson et al., 2004; An et al., 2005; Ward et al., 2005). The well-defined COI gene has demonstrated its efficacy as a dependable evolutionary marker (Baldwin et al., 1996;; An et al., 2005; Ward et al., 2005; Spies et al., 2006). The 12S ribosomal RNA (rRNA) gene sequences are extensively utilized in vertebrate phylogenetic analyses (Douzery and Catzeflis, 1995; Kjer, 1995; Lavergne et al., 1996; Ledje and Arnason, 1996; Richards and Moore, 1996; Springer and Douzery, 1996; Gatesy et al. 1997; Montgelard et al., 1997; Simons and Mayden, 1998). rRNA structures are more highly conserved among organisms than the sequences themselves (Springer and Douzery, 1996). It is, therefore, of particular importance that the structure is considered when rRNA genes are aligned, especially when the alignments are intended for phylogenetic analyses, because the phylogenetic inferences require comparisons of homologous characters of different sequences (Wang and Lee, 2002). Materials and MethodsEthical approvalOur study was approved by the research animal care and ethical committee of the Faculty of Science, Suez Canal University, under protocol REC241/2023. Samples collectionFish of the carangid species were collected from Egypt’s Red Sea (Fig. 1), Hurghada area, with the help of fishermen. The samples’ muscle tissues were taken from the caudal peduncle by using anatomy tools and kept in storage at −20°C for further use. Extraction of DNA and PCR amplificationThe entire genome’s DNA was obtained from the separated muscle tissues using the QIAamp DNA Mini kit (Qiagen, Hidden, Germany). We employed primers in accordance with the guidelines to amplify the COI gene as described by (Ward et al., 2005) in the five carangid fishes. However, the small mitochondrial rRNA (12S rRNA) gene was amplified using a forward primer in accordance with (Jin et al., 2013). The 25 μl of PCR master mix, 1 μl of each genomic DNA, forward and reverse primers, and a final reaction volume of 50 μl make up the PCR reactions. PCR cycling conditions included a 5-minute initial denaturation at 94°C, 30 cycles of 60-second denaturation at 94°C, 60-second annealing at 61°C (COI) and 50°C (12S rRNA), and an extension at 72°C for 60 seconds with post-cycling extension at 72°C for 7 minutes. The PCR products were separated using a 1.5% agarose gel stained with ethidium bromide. Sequencing and phylogenetic tree constructionMacrogen (Seoul, South Korea) performed all DNA sequencing using the same amplification primer. We submitted the sequences to the National Center for Biotechnology Information (GenBank/NCBI) of the COI gene and the 12S rRNA gene to assign accession numbers. We utilized MEGA version 11 to construct phylogenetic trees using the neighbor joining, minimum evolution, and maximum likelihood phylogenetic methodologies (Tamura et al., 2021), with bootstrap analysis conducted using 1,000 replicates (Felsenstein, 1985).
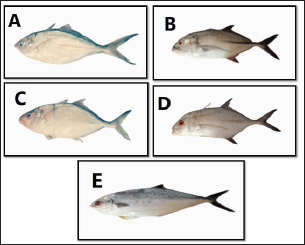
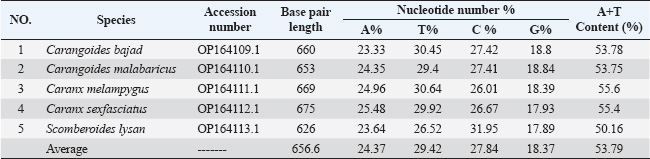
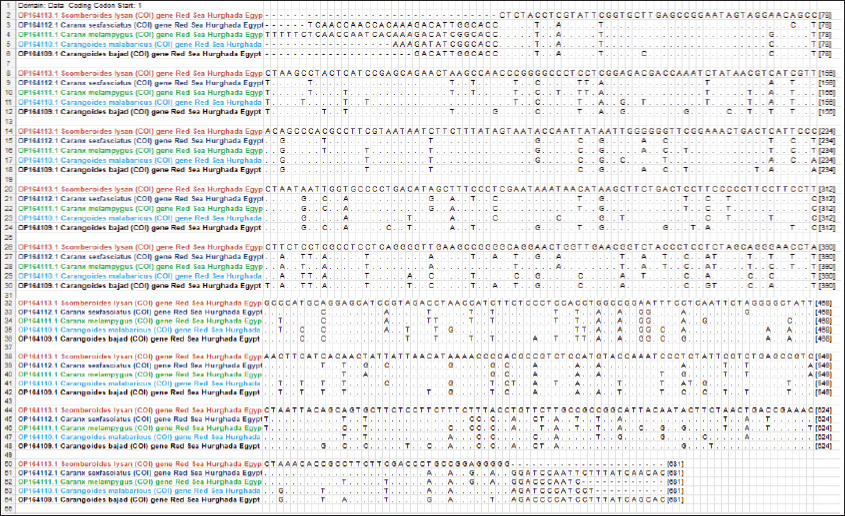
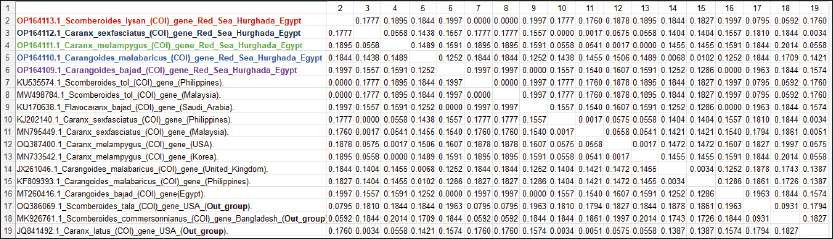
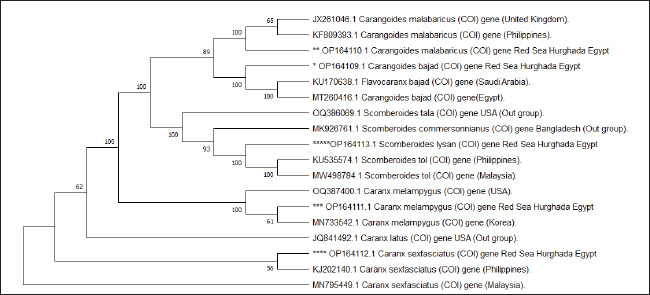
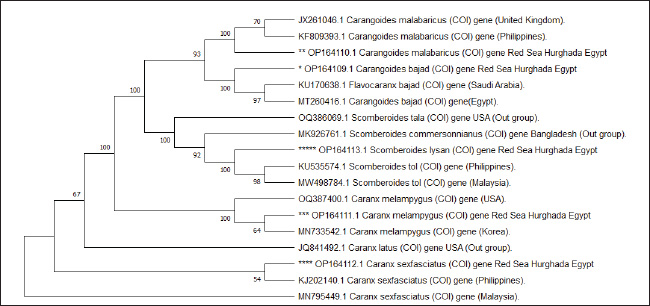
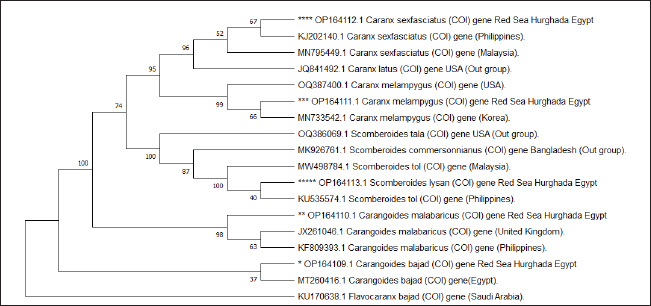
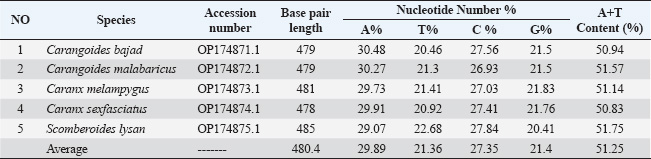
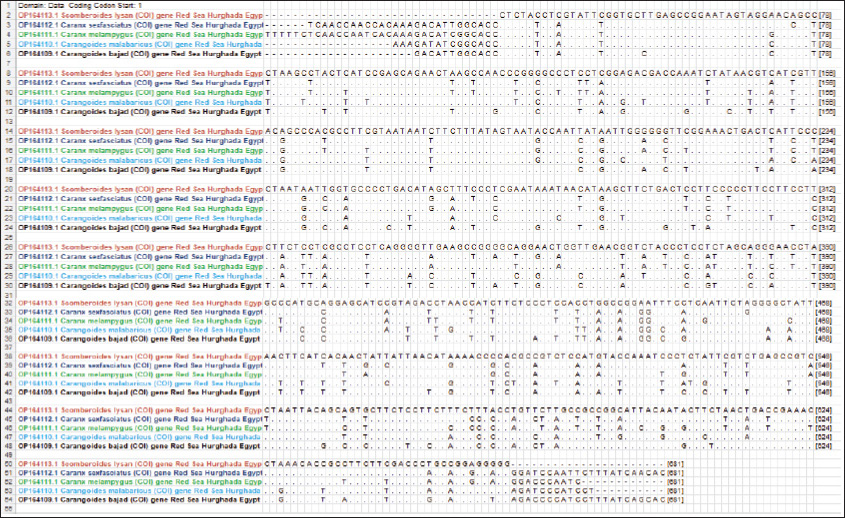
Fig. 1. Species of the Carangidae family, A-Carangoides bajad, B-Carangoides malabaricus, C-Caranx melampygus, D-Caranx sexfasciatus, E-Scomberoides lysan. ResultsThe length of cytochrome c oxidase subunit I (COI) gene sequences in five Carangid fishes expanded from 626 to 675 bp. The nucleotide sequences were inserted into the GenBank/NCBI with accession numbers (OP164109.1–OP164113.1). Our results showed that the longest nucleotide sequence (675 bp.) was found in Caranx sexfasciatus, while Scomberoides lysan showed the shortest sequence (626 bp.). The average frequencies of nucleotides were 24.37%, 29.42%, 27.84% and 18.37% for adenine (A), thymine (T), cytosine (C) and guanine (G), respectively. The COI gene displayed A+T ratio bigger than the C+G ratio in all species (Table 1). The average content of C+G ranged from 44.60% to 49.84 %, which was lower than the A+T in all species. The COI gene sequences from the five Carangid species showed that the final alignments had 681 bp; the conserved, parsimony informative, and variable sites were 494, 89, and 181, respectively (Fig. 2). MEGA version 11 was used to assess pairwise genetic distances between the outgroup of the COI gene and five carangid fishes (Fig. 3). For every Carangid species, the COI gene’s P-distances ranged from 0.0000 to 0.1997 percent. The maximum value (0.1997) was shown between Scomberoides_lysan_’Hurghada_Egypt’ (OP164113.1) and Flavocaranx_bajad_ ‘Saudi_Arabia’ (KU170638.1) and also between Carangoides_bajad ‘Hurghada_Egypt’ (OP164109.1) and Scomberoides_tol ‘Philippines’ (KU535574.1) and Scomberoides_tol ‘Malaysia’ (MW498784.1). However, 0.000 value was observed between Scomberoides_lysan_’Hurghada_Egypt’ (OP164113.1) and Scomberoides_tol ‘Philippines’ (KU535574.1) and Scomberoides_tol ‘Malaysia’ (MW498784.1) and also between Caranx_sexfasciatus ‘Hurghada_Egypt’ (OP164112.1), and Caranx_sexfasciatus ‘Philippines’ (KJ202140.1) and also between Caranx_melampygus ‘Hurghada_Egypt’ (OP164111.1) and Caranx_melampygus ‘Korea’ (MN733542.1). The P-distances among the understudied Carangid fish expanded from 0.0016% to 0.1997 %. The highest value (0.1997) was found between Scomberoides_lysan_’Hurghada_Egypt’ (OP164113.1) and Carangoides bajad ‘Egypt’ (MT260416.1). However, the lowest P-distance (0.0016) was found between Caranx_sexfasciatus ‘Hurghada_Egypt’ (OP164112.1) and Caranx_sexfasciatus ‘Malaysia’ (MN795449.1). To complete the phylogenetic tree investigation by the dint of cytochrome c oxidase subunit I (COI) gene sequence, the sequences acquired from five species of the family Carangidae were exercised in this work for a wide combination phylogenetic investigation. For widely illustrative phylogenetic investigation by using the cytochrome c oxidase subunit I (COI) gene, neighbor joining, minimum evolution, and maximum likelihood phylogenetic methods were used. With some variation in the support rate (Figs. 4 In five carangid fish, the length of the 12S rRNA gene sequence ranged from 478 to 485 bp. In five carangid fish, the length of the 12S rRNA gene sequence increased from 478 to 485 bp. With accession numbers, the nucleotide sequences were added to GenBank/NCBI (OP174871.1–OP174875.1). Our results showed that the longest nucleotide sequence (485 bp) was found in Caranx sexfasciatus, while Scomberoides lysan showed the shortest sequence (478 bp). Adenine (A), thymine (T), cytosine (C), and guanine (G) had average frequencies of 29.89%, 21.36%, 27.35%, and 21.4%, respectively. In every species, the A+T ratio was greater than the C+G ratio for the 12S rRNA gene (Table 2). C+G’s average content varied between 48.25 and 49.17. The 12S rRNA gene sequences from the five Carangid species found in Egypt’s Red Sea revealed the final alignments included 555 bp; the conserved, parsimony informative, and variable sites were 384, 24, and 95, respectively (Fig. 7). The sequences obtained from five species of the family Carangidae were used for a phylogenetic analysis based on the 12S rRNA gene, MEGA version 11 was used to assess pairwise genetic distances between the outgroup of 12S rRNA gene and five carangid fishes (Fig. 8). A comprehensive phylogenetic investigation was conducted using neighbor joining, minimum evolution, and maximum likelihood methods. The resulting trees showed some variation in support values (Figs. 9 Table 1. Accession number of the cytochrome c oxidase subunit I (COI) gene sequences in five Carangid fishes.
Fig. 2. Alignment of the COI gene sequences in five Carangid fishes. Dots show similar nucleotides while A, C, G, and T display the variance nucleotides.
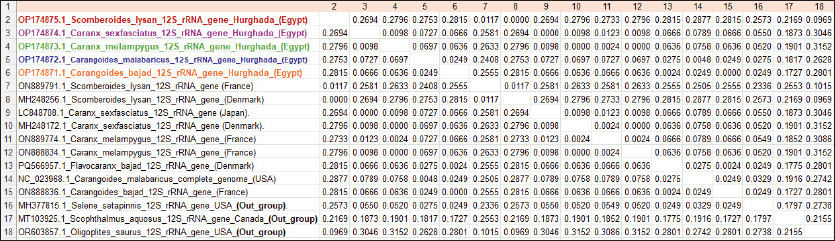
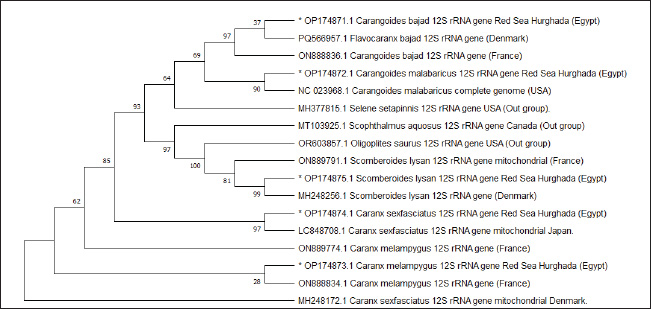
Fig. 3. Pairwise distances by the mean of the cytochrome c oxidase subunit I (COI) gene sequences in five Carangid fishes with their linkage species from the GenBank/NCBI. All sequence pairs’ unclear places were eliminated (pairwise deletion option). P-distances of the 12S rRNA gene for all the Carangid species ranged from 0.0000% to 0.28145%. The highest value (0.28145) was observed between Scomberoides_lysan ‘Hurghada_Egypt’ (OP174875.1) and Carangoides_malabaricus ‘USA’ (NC_023968.1). However, 0.000 value was observed between Scomberoides_lysan ‘Hurghada_Egypt’ (OP174875.1) and ‘Scomberoides_lysan’ ‘Denmark’ (MH248256.1) and between Caranx_sexfasciatus ‘Hurghada_Egypt’ (OP174874.1) and Caranx_sexfasciatus ‘Japan’ (LC848708.1). The P-distances among the understudied Carangid fish expanded from 0.00239% to 0.31524%. The highest value (0.31524) was found between Caranx_sexfasciatus ‘Denmark’ (MH248172.1) and Oligoplites_saurus ‘USA’ (Out_group) (OR603857.1). However, the lowest P-distance (0.00239) was found between Caranx_sexfasciatus ‘Denmark’ (MH248172.1) and Caranx_sexfasciatus ‘Hurghada_Egypt’ (OP174874.1). DiscussionCarangids are abundant and commercially important marine fish that contribute to a significant portion of the fisheries in many parts of the world (Jose et al., 2022). Efforts to use a segment of DNA as a barcode of identity have successfully employed COI to identify various taxa, including fish species (Ward et al., 2005). Ribosomal genes and its associated spacers are considered among the most versatile sequences for phylogenetic examinations (Hershkovitz and Lewis, 1996; Coleman, 2000; Coleman and Vacquier, 2002; Álvarez and Wendel, 2003; Coleman, 2003; Müller et al., 2007; Wickramasinghe et al., 2009; Yan et al., 2013). The sequences length of the cytochrome c oxidase subunit I (COI) gene sequences in five Carangid fishes expanded from 626 to 675 bp. which was in the expected length according to several studies (Lakra et al., 2011; Bingpeng et al., 2018; Ali et al., 2020; Allam and Welson, 2023). Adenine (A), thymine (T), cytosine (C), and guanine (G) had respective average frequencies of 24.37%, 29.42%, 27.84%, and 18.37% in the COI gene, according to the data. In every species, the COI gene showed an A+T ratio greater than the C+G ratio. This was supported by multiple studies. Bingpeng et al. (2018) revealed that during the examination of the DNA barcoding of various fish species in the Taiwan Strait, the mean AT content (53.20%) was greater than the GC content (46.80%). Alam et al. (2022) discovered that, when using the mitochondrial COI gene to analyze the genetic diversity of some Channa striata populations, the A+T content was higher than the G+C Rakh et al. (2023) revealed a high A+T content COI gene during a Bangladeshi population investigation.
Fig. 4. Neighbor-joining phylogenetic tree in five Carangid fishes and their outgroup using the COI gene.
Fig. 5. Minimum evolution phylogenetic tree in five Carangid fishes and their outgroup using the COI gene. The COI gene is a highly conserved region, which was extracted from five Carangid species from Egypt’s Red Sea and was in accordance with Ferrari et al. (2022), who applied COI barcoding to some fish samples of Amphiliidae and Cichlidae and found the percentage of conserved sites was 62.28%. The average frequencies of the 12S rRNA nucleotides were 29.89%, 21.36%, 27.35%, and 21.4 % for adenine (A), thymine (T), cytosine (C), and guanine (G), respectively. The 12S rRNA gene displayed A+T ratio greater than the C+G ratio in all species. This was agreed upon in many studies; Sivaraman et al. (2009) used 12S rRNA gene in four Cyprinid species. Widayanti et al. (2021) studied the genetic variation and phylogenetic analysis of Indonesian indigenous catfish (baung fish) based on mitochondrial 12S rRNA gene. Additionally, Wang et al. (2023) proclaim low G+C content of 12S rRNA in fish samples from Zhoushan coastal waters. The highly conserved sites of the 12S rRNA gene obtained from the five Carangid species from the Red Sea in Egypt, the fishes under study, agreed with various previous studies. Mahrous and Allam (2022) found similar findings during their study on catfish species using 12S rRNA gene. As well as Aziz et al. (2024) also found similar findings during their study phylogenetic relationship of some species of family Apogonidae using 12S rRNA.
Fig. 6. Maximum likelihood phylogenetic tree in five Carangid fishes and their outgroup using the COI gene. Table 2. Accession number, nucleotide frequencies, A+T contents, and their averages of 12S rRNA gene sequences in five carangid fishes.
Caranx exhibits physical similarities that make distinguishing it from other members of the Carangoides family challenging. Many authors grouped these species into the same genus as a result of these similarities (Smith-Vaniz, 1984; Reed et al., 2002). Some species form species complexes due to their wide geographic range and cryptic taxonomic characteristics (Smith-Vaniz and Carpenter, 2007). Mar’ie and Allam (2017) observed a robust relation between Caranx and Carangoides genera. Carangidae are highly valuable economic species that are significant to the ecology. The earlier investigations found that the evolutionary relationships in the genera of the subfamily Caranginae were quite complex; the number, connection, and each genus from each branch were agreed upon (Thu et al., 2019).. Numerous studies were conducted to evaluate the evolutionary relationship between species and genera in the Carangidae family using different DNA markers because of the complex position of the phylogenetic relationships between the genera in the Caranginae subfamily (Damerau et al., 2018; Torres and Santos, 2019; Li et al., 2020; Allam and Mar’ie, 2021; Jose et al., 2022). The sequences of the mitochondrial DNA, COI, and 12S rRNA genes were investigated to identify species and determine the evolutionary relationships between the Egyptian carangid fish species, which are very valuable as food and have a significant commercial impact. ConclusionAmong the world’s most economically significant coastal pelagic fishes are the Carangidae. There is taxonomic confusion among Carangid fishes due to morphological and meristic similarities. In this study, the small mitochondrial rRNA (12S rRNA) and the COI genes were used to access molecular diversity and conduct phylogenetic analysis of several Carangid species. The resulting phylogenetic analysis trees based on both genes provided insights into evolutionary relationships within the family of the Carangid species. Our results confirmed thermostability and environmental adaptation of the five species of the Carangidae family due to higher A+T content. Our results also confirmed the earlier conclusions of other authors that several genera of the Carangidae family are not monophyletic which does not include all the descendants of a common ancestor (Paraphyletic) and demonstrated the usefulness of the 12S rRNA gene and the COI gene in the phylogenetic analysis of the Carangid species.
Fig. 7. Alignment of (12S rRNA) sequences in five Carangid fishes. Dots show similar nucleotides while A, C, G, and T display the variance nucleotides.
Fig. 8. Pairwise distances by the mean of 12S rRNA gene amongst five species of Carangidae family with their linkage species from the GenBank/NCBI. AcknowledgmentsThe authors sincerely thank the administration of the Science, Technology & Innovation Funding Authority (STDF) for their support. The authors thank the generous funding with the Grant Post Graduate Support Grant (PGSG)- Call 2- PhD, Project ID: 49056, thanks for co-operation and efforts for supporting scientific research in Egypt. Conflict of interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. FundingThis study is based on research that was funded by Science, Technology and Innovation Funding Authority (STDF) under grant “Post Graduate Support Grant (PGSG)- Call 2- PhD. Project ID: 49056. Authors’ contributionsSara Gamal A. Eisa: Methodology, Data Curation, Investigation, and Writing; Mohammad Allam: Conceptualization, Data Curation, Review & Editing; Ali Hussein Abu Almaaty: Investigation, and Writing - Review & Editing; Mohamed Kamel Hassan: Data Curation & Editing. Data availability
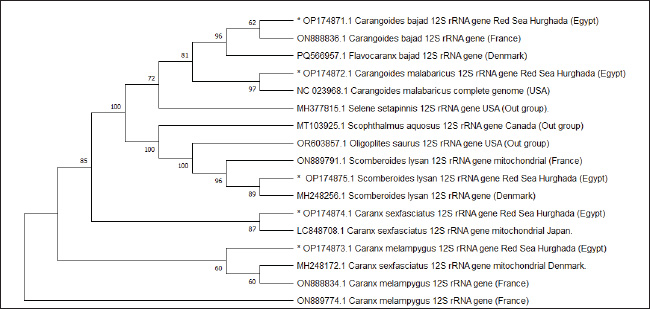
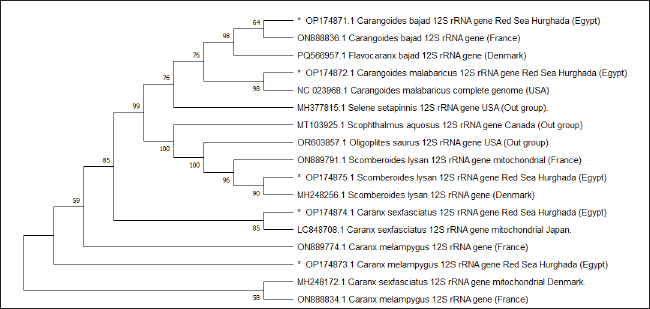
Fig. 9. Neighbor joining phylogenetic tree in five Carangid fishes and their outgroup using the 12S rRNA gene.
Fig. 10. Minimum evolution phylogenetic tree in five Carangid fishes and their outgroup using the 12S rRNA gene
Fig. 11. Maximum likelihood phylogenetic tree in five Carangid fishes and their outgroup using the 12S rRNA gene ReferencesAlam, M.S., Projna, F., Zafrin, M.S., Das, R. and Khan, M.G.Q. 2022. Assessment of genetic diversity, detection of strain-specific single nucleotide polymorphisms, and identification of the Bangladesh and Vietnam strain of Channa striata by PCR-RFLP analysis of the mitochondrial CO1 gene fragment. Aqua. Fish. 7(3), 287–295. Allam M. and Welson, M. 2023. DNA barcoding of some species of genus synodontis (Family Mochokidae) from the River Nile in Egypt. Egypt. J. Aquatic Biol. Fisheries 27, 199–210. Ali, F.S., Ismail, M. and Aly, W. 2020. DNA barcoding to characterize biodiversity of freshwater fishes of Egypt. Mol. Biol. Rep. 47(6), 5865–5877. Ali, F.S., Mamoon, A. and Abbas, E. 2021. Mitochondrial-based phylogenetic inference of worldwide species of genus Siganus. Egypt. J. Aquatic. Biol. Fish. 25, 371–388; doi:10.21608/ejabf.2021.143930 Allam, M. and Marie, Z.A. 2021. Phylogenetic and genetic diversity of some carangid species from the Egyptian red sea using divergent domain D11 Of 28S rRNA gene. Egypt. J. Aquatic Biol. Fisheries 25(1), 61–73. Alvarez, I. and Wendel, J.F. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29(3), 417–434. An, H.S., Jee, Y.J., Min, K.S., Kim, B.L. and Han, S.J. 2005. Phylogenetic analysis of six pacific abalone (Haiotidae) based on DNA sequences of 16S rRNA and cytochrome c oxidase subunit I mitochondrial genes. Mar. Biol. 7, 373–380. Aziz, M.M., Abu Almaaty, A.H. and Allam, M. 2024. Phylogenetic inference of some species of the family apogonidae using 12S rRNA sequence. Egypt. J. Aquatic Biol. Fish. 28(4), 55–65. Baldwin, B.S., Black, M., Sanjur, O., Gustafson, R., Lutz. R.A. and Vrijenhoek, R.C. 1996. A diagnostic molecular marker for zebra mussels (Dreissena polymorpha) and potentially co-occurring bivalves: mitochondrial COI. Mol. Mar. Biol. Biotechnol. 5, 9–14. Bermingham, E. and Lessios, H.A. 1993. Rate variation of protein and mitochondrial DNA evolution as revealed by sea urchins separated by the Isthmus of Panama. Proc. Nat. Acad. Sci. 90(7), 2734–2738. Bingpeng, X., Heshan, L., Zhilan, Z., Chunguang, W., Yanguo, W. and Jianjun, W. 2018. DNA barcoding for identification of fish species in the Taiwan Strait. PLoS One 13(6), e0198109. Birstein, V.J. and Desalle, R. 1998. Molecular phylogeny of acipenserinae. Mol. Phylogenet. Evol. 9(1), 141–155. Bohlke, J.E. and Chaplin, C.C.G. 1993. Fishes of the Bahamas and adjacent tropical water. A review of the biology of the family carangidae with emphasis on species found Hawaiian waters. Ed., Honebrink, R.R. DAR. Tech. Report 20-01. Brown, J.R., Beckenbach, K., Beckenbach, A.T. and Smith, M.J. 1996. Length variation, heteroplasmy and sequence divergence in the mitochondrial DNA of four species of sturgeon (Acipenser). Genetics 142, 525–535. Brown, W.M 1985. The mitochondrial genome of animals: molecular evolutionary genetics. In Molecular evolutionary genetics. New York: Plenum Press, pp 95–130. Burgener, M. and Hubner, P. 1998. Mitochondrial DNA enrichment for species identification and evolutionary analysis. Z. Lebensm. Unters. Forsch. 207, 261–263. Cano, R.J., Poinar, H.N., Pieniazek, N.J., Acra, A. and Poinar, G.O. 1993. Amplification and sequencing of DNA from a 120–135-million-year-old weevil. Nature 363, 536–538. Chow, S., Clarke, M.E. and Walsh, P.J. 1993. PCR-RFLP analysis on thirteen western Atlantic snappers (subfamily Lutjaninae): a simple method for species and stock identification. Fish. Bull. 91, 619–627. Coleman, A.W. 2000. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 151(1), 1–9. Coleman, A.W. 2003. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 19(7), 370–375. Coleman, A.W. and Vacquier, V.D. 2002. Exploring the phylogenetic utility of ITS sequences for animals: a test case for abalone (Haliotis). J. Mol. Evol. 54(2), 246–257. Collura, R.V. and Stewart, C.B. 1995. Insertions and duplications of mtDNA in the nuclear genomes of Old World monkeys and hominoids. Nature 378, 485–489. Damerau, M., Freese, M. and Hanel, R. 2018. Multi-gene phylogeny of jacks and pompanos (Carangidae), including placement of monotypic vadigo Campogramma glaycos. J. Fish. Biol. 92(1), 190–202. DeSalle, R., Williams, A.K. and George, M. 1993. Isolation and characterization of animal mitochondrial DNA. Methods Enzymol. 224, 176–204. Douzery, E. and Catzeflis, F.M. 1995. Molecular evolution of the mitochondrial 12S rRNA in Ungulata (mammalia). J. Mol. Evol. 41, 622–636. Esposti, M.D., De Vries, S., Crimi, M., Ghelli, A., Patarnello, T. and Meyer, A. 1993. Mitochondrial cytochrome b: evolution and structure of the protein. Biochim. Biophys. Acta. 1143, 243–271. Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4), 783–791. Ferrari, C., Tovela, E., Taviani, E. and Nonnis Marzano, F. 2022. DNA barcoding to assess species identification in museum samples of Amphiliidae and natural samples of Cichlidae from Southern Mozambique. Rend. Fis. Acc. Lincei. 33(4), 713–720. G. K.Garcia-Morales, A.E. and Elias-Gutierrez, M. 2013. DNA barcoding freshwater Rotifera of Mexico: evidence of cryptic speciation in common rotifers. Mol. Ecol. Resour. 13(6), 1097–1107. Gatesy, J., Mato, G.A., Vrba, E., Schaler, G. and Desalle, R. 1997. A cladistic analysis of mitochondrial ribosomal DNA from Bovidae. Mol. Phylogenet. Evol. 7, 303–319. Gushiken, S. 1988. Phylogenetic relationships of the perciform genera of the family Carangidae. Ichthyol. Res. 34(4), 443–461. Hayashi, J.I., Tagashira, Y. and Yoshida, M.C. 1985. Absence of extensive recombination between inter- and intraspecies mitochondrial DNA in mammalian cells. Exp. Cell Res. 160, 387–395. Hershkovitz, M.A. and Lewis, L.A. 1996. Deep level diagnostic value of the rDNA-ITS region: the case of an algal interloper. Mol. Biol. Evol. 13(9), 167–177. Holland, B.S., Dawson, M.N., Crow, G.L. and Hofmann, D.K. 2004. Global phylogeography of Cassiopea (Scyphozoa: rhizostomeae): molecular evidence for cryptic species and multiple invasions of the Hawaiian Islands. Mar. Biol. 145, 1119–1128. Honebrink, R. 2000. A review of the biology of the family Carangidae, with emphasis on species found in Hawaiian waters. Irwin, D.M., Kocher, T.D. and Wilson, A.C. 1991. Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32, 128–144. Jaafar, T.N., Taylor, M.I., Mohd Nor, S.A., de Bruyn, M. and Carvalho, G.R. 2012. DNA barcoding reveals cryptic diversity within commercially exploited Indo-Malay Carangidae (Teleostei: perciformes). PLoS One 7(11), 49623. Jawad, L.A. and Al-Mamry, J.M. 2012. Relationship between fish length and otolith dimensions in the Carangid Fish (Carangoides coeruleopinnatus (Rüppell, 1830)) collected from the Sea of Oman. J. Fish. Sci. 6(3), 203–208. Jin, X.X., Zhao, S.L. and Wang, R.X. 2013. Universal primers to amplify the complete mitochondrial 12S rRNA gene in marine fish species. Gene Mol. Res. 12(4), 4575–4578. Jose, A., Sukumaran, S., Mukundan, L.P., Raj, N., Mary, S., Nisha, K. and Gopalakrishnan, A. 2022. Comparative mitogenomics and phylogenetics of the family Carangidae with special emphasis on the mitogenome of the Indian Scad Decapterus russelli. Sci. Rep. 12(1), 5642. Kjer, K.M. 1995. Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs. Mol. Phylogenet. Evol. 4, 314–330. Lakra, W.S., Verma, M.S., Goswami, M., Lal, K.K., Mohindra, V., Punia, P., Gopalakrishnan, A., Singh, K.V., Ward, R.D. and Hebert, P. 2011. DNA barcoding Indian marine fishes. Mol. Ecol. Resour. 11(1), 60–71. Laroche, W.A., Smith-Vaniz, W.F. and Richardson, S.L. 1984. Carangidae: development in ontogeny and systematics of fishes. Am. Soc. Ichthyol. Herpetol. 510–522. Lavergne, A., Douzery, E., Stichler, T., Catzeflis, F.M. and Springer, M.S. 1996. Interordinal mammalian relationships: evidence for Paenungulate Monophyly is provided by complete mitochondrial 12S rRNA sequences. Mol. Phylogenet. Evol. 6, 245–258. Le Roux, J. and Wieczorek, A.M. 2009. Molecular systematics and population genetics of biological invasions: towards a better understanding of invasive species management. Ann. Appl. Biol. 154(1), 1–17. Ledje, C. and Arnason, U. 1996. Phylogenetic relationships within Caniform carnivores based on analysis of the mitochondrial 12S rRNA gene. J. Mol. Evol. 43, 641–649. Li, Z., Li, M., Xu, S., Liu, L., Chen, Z. and Zou, K. 2020. Complete mitogenomes of three carangidae (perciformes) fishes: genome description and phylogenetic considerations. Int. J. Mol. Sci. 21(13), 4685. Lin. P.-L., Shao. K.-T. 1999. A review of the carangid fishes (Family Carangidae) from Taiwan with descriptions of four new records. Zool. Stud. Taiwan 38, 33–68. Mahrous, N.S. and Allam, M. 2022. Phylogenetic relationships among some catfishes assessed by small and large mitochondrial rRNA sequences. Egypt. J. Aquatic. Biol. Fish. 26(6), 1069–1082. Mar’Ie, Z.A. and Allam, M. 2017. Using start codon targeted (SCoT) polymorphism for genetic diversity analysis of three Red Sea fishes (Family: carangidae). IOSR J. Pharm. Biol. Sci. 12(1), 50–56. Montgelard, C., Catzeflis, F.M. and Douzery, E. 1997. Phylogenetic relationships of artiodactyls and cetaceans as deduced from the comparison of cytochrome b and 12S rRNA mitochondrial sequences. Mol. Biol. Evol. 14, 550–559. Müller, T., Philippi, N., Dandekar, T., Schultz, J., Wolf, M. 2007. Distinguishing species. RNA 13(9), 1469–1472. Munasinghe, D.H.N., Burridge, C.P. and Austin, C.M. 2004. The systematics of freshwater crayfish of the genus Cherax Erichson (Decapoda: parastacidae) in eastern Australia re-examined using nucleotide sequences from 12S rRNA and 16S rRNA genes. Invertebr. Syst. 18(2), 215–225. Rakhi. R.F. 2023. Investigating the population genetic structure of the endangered great snakehead (Channa marulius) in open waterbodies of Bangladesh using mitochondrial DNA markers. Egypt. J. Aquatic Biol. Fisheries 27(3), 979–995. Randall, JE. 1995. Coastal fishes of Oman. New South Wales, Australia: Crawford House Publishing Pty Ltd, Bathurst, pp: 439. Reed, D.L., Carrpenter, K.E. and deGravelle, M.J. 2002. Molecular systematic of the Jacks (Perciformes: carangidae) based on mitochondrial cytochrome b sequences using parsimony, likelihood, and Bayesian approaches. Mol. Phylogenet. Evol. 23(3), 513–524. Richards, C.M. and Moore, W.S. 1996. A phylogeny for the African treefrog family hyperoliidae based on mitochondrial rDNA. Mol. Phylogenet. Evol. 5, 522–532. Sivaraman, G.K., Barat, A. and Mahanta, P.C. 2009. Molecular phylogeny of cyprinid fishes of India using 12S rRNA gene sequences. Environ. Ecol 4, 43–53. Santos, S., Schneider, H. and Sampaio, I. 2003. Genetic differentiation of Macrodon ancylodon (Sciaenidae, Perciformes) populations in Atlantic coastal waters of South America as revealed by mtDNA analysis. Gene Biol. Mol. 26, 151–161. Simons, A.M. and Mayden, R.L. 1998. Phylogenetic relationships of the Western North American Phoxinins (Actinopterygii: cyprinidae) as inferred from mitochondrial 12S and 16S ribosomal RNA sequences. Mol. Phylogenet. Evol. 9, 308–329. Smith-Vaniz, W.F. 1984. Carangidae: relationships. Am. Soc. Ichthyol. Herpetol. 1, 640–670. Smith-Vaniz, W.F. and Carpenter, K.E. 2007. Review of the crevalle jacks, Caranx hippo complex (Teleostei: carangidae), with a description of a new species from West Africa. Fish. Bull. 105(2), 207–233. Spies, I.B., Gaichas, S., Stevenson, D.E., Orr, J.W. and Canino, M.F. 2006. DNA based identification of Alaska skates (Amblyraja, Bathyraja and Raja: rajidae) using cytochrome c oxidase submit (COI) variation. J. Fish. Bull. 69(B), 283–292. Springer, M.S. and Douzery, E. 1996. Secondary structure and patterns of evolution among Mammalian mitochondrial 12S rRNA molecules. J. Mol. Evol. 43, 357–373. Tamura, K., Stecher, G. and Kumar, S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38(7), 3022–3027. Thu, P., Linh, N.M, Van Quan, N., Van Chien, P., Ly, D.H. and Hiep, L.B. 2019. DNA barcoding for identification of some fish species (Carangidae) in Vietnam coastal area. Vietnamese J. Mar. Sci. Technol. 19(4), 527–536. Torres, S.K.M. and Santos, B.S. 2019. Species identification among morphologically-similar caranx species. Turkish J. Fish. Aquatic Sci. 20(2), 159–169. Vinson, C., Grazielle, G., Schneider, H. and Sampaio, I. 2004. Sciaenidae fish of the Caete river estuary, Northern Brazil: mitochondrial DNA suggests explosive radiation for the Western Atlantic assemblage. Genet. Mol. Biol. 27(2), 174–180. Wang, H. and Lee, S. 2002. Secondary structure of mitochondrial 12S rRNA among fish and its phylogenetic applications. Mol. Biol. Evol. 19(2), 138–148. Wang, Y., Song, N., Liu, S., Chen, Z., Xu, A. and Gao, T. 2023. DNA barcoding of fishes from Zhoushan coastal waters using mitochondrial COI and 12S rRNA genes. J. Oceanol. Limnol. 41(5), 1997–2009. Ward, R.D., Zemlak, T.S., Innes, B.H., Last, P.R. and Hebert, P.D. 2005. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360(1462), 1847–1857. Wickramasinghe, S., Yatawara, L., Rajapakse, R.P.V.J., Agatsuma, T. 2009. Toxocara canis and Toxocara vitulorum: molecular characterization, discrimination, and phylogenetic analysis based on mitochondrial (ATP synthase subunit 6 and 12S) and nuclear ribosomal (ITS-2 and 28S) genes. Parasitol. Res. 104(6), 1425–1430. Widayanti, R., Kusumaastuti, K.A., Novi, J.M., Adani, F.K., Gultom, C.R.P., Prastiti, A.D., Nugroho, H.A. and Pakpahan, S. 2021. Genetic variation and phylogenetic analysis of Indonesian indigenous catfish (baung fish) based on mitochondrial 12S rRNA gene. Vet. World 14(3), 751–757; doi:10.14202/vetworld.2021.751-757 Yan, H., Lou, Z., Li, L., Ni, X., Guo, A., Li, H., Zheng, Y., Dyachenko, V. and Jia, W. 2013. The nuclear 18S ribosomal RNA gene as a source of phylogenetic information in the genus Taenia. Parasitol. Res. 112(3), 1343–1347. Yang, L., Tan, Z., Wang, D., Xue, L., Guan, M.X., Huang, T. and Li, R. 2014. Species identification through mitochondrial rRNA genetic analysis. Sci. Rep. 4(1), 408. | ||
| How to Cite this Article |
| Pubmed Style Allam M, Eisa SGAA, Almaaty AHA, Hassan MK. Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes. Open Vet. J.. 2025; 15(8): 3809-3822. doi:10.5455/OVJ.2025.v15.i8.46 Web Style Allam M, Eisa SGAA, Almaaty AHA, Hassan MK. Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes. https://www.openveterinaryjournal.com/?mno=245625 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i8.46 AMA (American Medical Association) Style Allam M, Eisa SGAA, Almaaty AHA, Hassan MK. Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes. Open Vet. J.. 2025; 15(8): 3809-3822. doi:10.5455/OVJ.2025.v15.i8.46 Vancouver/ICMJE Style Allam M, Eisa SGAA, Almaaty AHA, Hassan MK. Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes. Open Vet. J.. (2025), [cited January 25, 2026]; 15(8): 3809-3822. doi:10.5455/OVJ.2025.v15.i8.46 Harvard Style Allam, M., Eisa, . S. G. A. A., Almaaty, . A. H. A. & Hassan, . M. K. (2025) Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes. Open Vet. J., 15 (8), 3809-3822. doi:10.5455/OVJ.2025.v15.i8.46 Turabian Style Allam, Mohammad, Sara Gamal Abd Allah Eisa, Ali Hussein Abu Almaaty, and Mohamed Kamel Hassan. 2025. Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes. Open Veterinary Journal, 15 (8), 3809-3822. doi:10.5455/OVJ.2025.v15.i8.46 Chicago Style Allam, Mohammad, Sara Gamal Abd Allah Eisa, Ali Hussein Abu Almaaty, and Mohamed Kamel Hassan. "Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes." Open Veterinary Journal 15 (2025), 3809-3822. doi:10.5455/OVJ.2025.v15.i8.46 MLA (The Modern Language Association) Style Allam, Mohammad, Sara Gamal Abd Allah Eisa, Ali Hussein Abu Almaaty, and Mohamed Kamel Hassan. "Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes." Open Veterinary Journal 15.8 (2025), 3809-3822. Print. doi:10.5455/OVJ.2025.v15.i8.46 APA (American Psychological Association) Style Allam, M., Eisa, . S. G. A. A., Almaaty, . A. H. A. & Hassan, . M. K. (2025) Molecular phylogeny of some Carangid species from the Egyptian Red Sea using cytochrome c oxidase subunit I (COI) and small (12S rRNA) mitochondrial rRNA genes. Open Veterinary Journal, 15 (8), 3809-3822. doi:10.5455/OVJ.2025.v15.i8.46 |