| Review Article | ||
Open Vet. J.. 2025; 15(5): 1866-1879 Open Veterinary Journal, (2025), Vol. 15(5): 1866-1879 Review Article The global burden of swine influenza and its mitigationDewa Ketut Meles1, Aswin Rafif Khairullah2, Rimayanti Rimayanti3, Imam Mustofa3*, Wurlina Wurlina3, Suzanita Utama3, Tita Damayanti Lestari3, Sri Mulyati3, Riza Zainuddin Ahmad2, Ikechukwu Benjamin Moses4, Syahputra Wibowo5, Muhammad Khaliim Jati Kusala2, Bantari Wisynu Kusuma Wardhani6, Ima Fauziah2, Dea Anita Ariani Kurniasih7, Lili Anggraini8, Fitrine Ekawasti2 and Adeyinka Oye Akintunde91Division of Basic Veterinary Medicine, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 2Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia 3Division of Veterinary Reproduction, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 4Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, Nigeria 5Eijkman Research Center for Molecular Biology, National Research and Innovation Agency (BRIN), Bogor, Indonesia 6Research Center for Pharmaceutical Ingredients and Traditional Medicine, National Research and Innovation Agency (BRIN), Bogor, Indonesia 7Research Center for Public Health and Nutrition, National Research and Innovation Agency (BRIN), Bogor, Indonesia 8Research Center for Animal Husbandry, National Research and Innovation Agency (BRIN), Bogor, Indonesia 9Department of Agriculture and Industrial Technology, Babcock University, Ilishan-Remo, Nigeria *Corresponding Author: Imam Mustofa. Division of Veterinary Reproduction, Faculty of Veterinary Medicine, Universitas Airlangga, Jl. Dr. Ir. H. Soekarno, Kampus C Mulyorejo, Surabaya 60115, East Java, Indonesia. Email: imam.mustofa [at] fkh.unair.ac.id Submitted: 12/01/2025 Revised: 25/03/2025 Accepted: 20/04/2025 Published: 31/05/2025 © 2025 Open Veterinary Journal
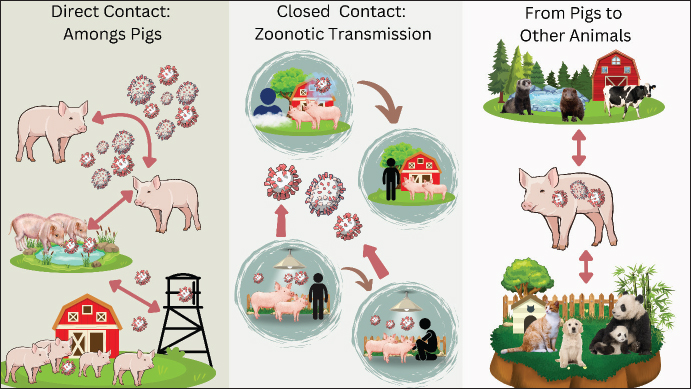
ABSTRACTSwine influenza, often known as swine flu, is a respiratory disease caused by type A influenza virus (IAV) called swine influenza virus (SIV). There are currently multiple subtypes of IAV in pigs, including H1N1, H1N2, and H3N2. While the other subtypes of IAV were only detected in pigs, the H1N1 strain was isolated from infected people. The process of SIV infection is similar to that of other respiratory viral infections: the virus enters the body through aerosol, and the infection spreads quickly to the nasal cavity and epithelium of major airways. Immune responses such as innate, mucosal, and systemic immunity (both humoral and cellular immunity) are triggered by IAV infection. SIVs, like the 2009 H1N1 pandemic strain, can be easily transmitted from pigs to humans, thereby causing significant public health concerns. People who contract new swine influenza infections have bexperienceiety of symptoms that resemble those of seasonal influenza. Pandemics like the 2009 H1N1 pandemic have substantial economic impacts due to the costs associated with prevention, treatment, and hospitalization. The 2009 H1N1 pandemic, a new strain of the H1N1 virus, spread rapidly to over 200 countries, causing an estimated 284,400 deaths worldwide, according to the World Health Organization. The primary symptoms are fever, chills, headache, runny nose, body aches, joint pain or myalgia, cough, sore throat, and exhaustion. The hemagglutinin sequence of SIVs is the primary basis for the development of polymerase chain reaction tests. In mammals, influenza viruses are spread by direct or indirect contact with nasal secretions, as well as by droplets and aerosols released during coughing and sneezing. Swine influenza most commonly attacks children aged 5 years and over and teenagers. This illness is treated with antibiotics, which help prevent bacterial pneumonia and other secondary illnesses in calves weakened by influenza. There is now an injectable vaccine for influenza A. Wholistic preventive approach and appropriate biosafety measures are important strategies for preventing the occurrence of viruses. Keywords: IAV, pig, public health, SIV, virus. IntroductionSwine influenza, often known as swine flu, is a respiratory disease caused by type A influenza virus (IAV) called swine influenza virus (SIV) (Ohanu et al., 2019). This virus belongs to the Orthomyxoviridae family and infects the respiratory tract of pigs (Richard and Fouchier, 2016). Although swine influenza sickness has a low fatality rate and high morbidity rate, it can produce more severe outbreaks, often leading to reduced piglet growth rates and associated financial losses (Ma, 2020). Occasionally, SIV infects humans and other species. The symptoms of swine influenza are similar to those of human influenza. Clinical instances in humans are often mild and mimic human influenza, although fatalities have been reported (Schnitzler and Schnitzler, 2009). Since 2009, swine influenza has been a serious health concern after the World Health Organization (WHO) declared it a pandemic (Dandagi and Byahatti, 2011). Numerous SIV variants have surfaced over time, and there are currently multiple main IAV subtypes in pigs, including H1N1, H1N2, and H3N2 (Huang et al., 2024). The other subtypes of IAV were detected only in pigs, and the H1N1 strain was isolated from infected people (Schnitzler and Schnitzler, 2009). The name of the enveloped RNA virus known as the SIV is derived from two surface antigens: N1 (neuramidase type 1) and H1 [hemagglutinin (HA) type 1] (Dou et al., 2018). It should be noted that although the 2009 pandemic H1N1 virus evolved in pigs, it was not derived from pigs. The virus is a quadruple-reassortant containing genes from human and avian influenza viruses, as well as genes from SIVs from North America, Europe, and Asia (Mena et al., 2016). Since its initial isolation from pigs in the United States in the 1930s, the most vulnerable groups for SIV infections have been veterinarians and pork producers (Lorusso et al., 2013). Humans are susceptible to this pathogenic virus, which manifests directly as decreased appetite, nasal discharge, and barking cough (Bhatta et al., 2020). The swine influenza became a pandemic because of its rapid spread and unchecked illnesses. In small children and the elderly, severe instances of swine influenza can result in pneumonia and even death (Cunha, 2010). Although the virus can spread among pigs all year round, swine influenza outbreaks often occur in the fall and winter, just like in people (Schmidt, 2009). In the past ten years, swine influenza has become one of the most deadly infectious diseases, spreading like a pandemic or epidemic all across the world. Today, this illness spreads all across the world as a human seasonal influenza virus (Ma, 2020). It has been passed from humans to pigs, where it has combined with other SIVs (Mo et al., 2022). Pig vaccination campaigns have become more challenging due to the increased virus diversity brought about by the occurrence of cross-transmission from humans to pigs and vice versa, as well as other modifications in the circulation of SIVs (Richt et al., 2006). The aim of this review article is to provide a comprehensive literature on swine influenza. The management of this illness depends on a thorough comprehension of the processes governing pathogenicity and interspecies transmission, as well as the availability of efficient therapeutic and preventive treatments. EtiologyInfluenza viruses are negative-sense RNA viruses belonging to the three genera Influenza A, B, and C of the Alphainfluenzavirus genus and the Orthomyxoviridae family (Bouvier and Palese, 2008). Influenza viruses are classified into subtypes based on two surface proteins: (HA) and neuraminidase (NA). For instance, subtype H1N2 is produced by a virus that possesses both type 1 HA and type 2 NA (Kosik and Yewdell, 2019). There is typically little to no cross-protection between different forms of HA or NA, and these two proteins are important targets of the immune response. Nine NAs (N1 to N9) and at least sixteen HA types (H1 to H16) are known to exist in birds. Two more HA and NA subtypes were found in bats, and a few avian subtypes were found in other mammals (Herfst et al., 2020). The influenza A virus species, which includes the SIV, is a type of virus that can infect humans, dogs, horses, and poultry (Short et al., 2015). The term “antigenic shift” refers to the slow alteration of the virus’s HA and NA genes caused by mutations. The current host immune response to the virus might no longer be effective if the HA and NA proteins are altered sufficiently (van de Sandt et al., 2012). The three primary influenza A virus subtypes that are most prevalent globally in pigs are H1N1, H1N2, and H3N2 (Anderson et al., 2021). H3N2 developed from H2N2 by means of an antigenic shift (Allen and Ross, 2018). The virus’s genes are constantly undergoing point mutations that result in antigenic changes, which assist the virus elude host defenses and lead to the emergence of new strains each year. The H1N1 strain of the SIVs, which is currently in circulation, has undergone three rounds of re-sequencing and incorporates genes from human, swine, and avian viruses (Hennig et al., 2022). Type B and C influenza viruses are not subtypes. There is evidence from serology and virology that pigs can occasionally contract human influenza B and C viruses (Brown et al., 1995). The type of surface, the surrounding environment, and the presence of organic material (such as feces) can all affect how long influenza viruses can survive in the environment. Protection from sunlight and cold temperatures boosts the survival of viruses. In untreated pork slurry, the SIV is rendered inactive after 1–2.5 hours at 50°C–55°C (122°F–131°F), 2 weeks at 20°C (68°F), and 9 weeks at 5°C (41°F) (Li and Robertson, 2021). It probably persists in water or on fomites, similar to other influenza viruses that infect mammals. On most surfaces, human IAVs survive for less than 24 to 48 hours, and in many settings, they frequently seem contagious for minutes to hours (Peteranderl et al., 2016). However, in rare circumstances, the virus may live longer. Avian influenza viruses and human IAVs can live for weeks or months in some types of water (such as distilled water), according to several laboratory tests (Poulson et al., 2016). However, they may be inactivated more quickly in aquatic habitats with normal microbial flora. HistorySwine influenza is identical to human influenza and was first proposed as a disease during the 1918 Spanish influenza epidemic when people and pigs contracted the same illness (Nelson and Worobey, 2018). The H1H1N1as the main cause of the 1918 influenza pandemic (Anhlan et al., 2011). In 1930, SIVs were initially isolated from pigs in the United States. That year, the prototype classical SIH1N1 strain (A/Swine/Iowa/30) was transferred to another pig for experimental examination and characterization, and swine influenza was first isolated in a pig (Brockwell-Staats et al., 2009). IAV invaded other parts of the world in the 1950s to 1970s, including the Hong Kong flu in 1968 and the Asian flu in 1957 (Lycett et al., 2019). Pigs were shown to be susceptible to avian and human influenza (H3N2) influenza in 1970, and Russia experienced a historic swine influenza outbreak in 1977 (Horimoto and Kawaoka, 2001). Three distinct subtypes and five distinct genotypes emerged as novel strains between 1997 and 2002, which were the primary causes of severe influenza in pigs in North America (Webster and Govorkova, 2014). First identified in two Mexican youngsters in 2009, H1N1 quickly spread around the globe (Hsieh et al., 2011). Owing to several deaths globally that year, the WHO upgraded the pandemic alert to phase 6 in relation to the novel H1N1 influenza virus (Dandagi and Byahatti, 2011). The 2009 pandemic infection was caused by the swine H1N1 virus, avian H1N1 virus, and human H3N2 virus. Three gene reassortments in the pig host, a combination of gene changes from three viruses with distinct genotypes, led to the formation of the novel H1N1 influenza virus (Mena et al., 2016). EpidemiologyThe H1N1 and H3N2 IAV subtypes have been extensively documented in pigs and are frequently linked to clinical illness (Anderson et al., 2021). These viruses include human and avian H3N2 viruses, as well as traditional swine H1N1 and avian H1N1 viruses. These viruses are among the most prevalent respiratory illnesses in pigs and are still mostly endemic in pig populations around the world (Peiris et al., 2001). According to the UK’s sero-surveillance statistics, over half of the country’s adult pig population has contracted one or more IAV in their lifespan, with 14% of pigs having contracted influenza viruses of both human and swine origins (Mastin et al., 2011). During the 1918 influenza pandemic, pigs contracted swine influenza concurrently with humans, leading to the first suggestion that the two diseases were connected (Nelson and Worobey, 2018). Reports of this illness have come from parts of Asia (Zeller et al., 2023), Africa (Anjorin et al., 2023), Europe (Henritzi et al., 2020), North America (Jhung et al., 2011), and South America (Cappuccio et al., 2011). SIV is considered enzootic in most locations with dense pig populations; however, the virus may go unnoticed in some areas because infected herds may be asymptomatic or have only moderate clinical indications (Ryt-Hansen et al., 2019). For older than 70 years, the most common virus among North American pigs has been classic H1N1 SIV (Gray et al., 2007). During this period, several human-acquired H3 viruses were detected in trace amounts, but they failed to form stable lineages in pigs. In the late 1990s, the triple-reassorted H3N2 virus initially appeared in North American swine, mostly in the Midwest of the United States, and then expanded to other areas (Webby et al., 2000). In North America, swine influenza is mostly caused by re-emerging H3N2 viruses from humans, pigs, and birds. H1N1 and H3N2 have resorted to one another, resulting in H1N2 (Bhatta et al., 2020). After several cases of SIV spread throughout Europe, the H1N1 virus, which originated in birds, infiltrated the European pig population in the late 1970s (Ma, 2020). In pigs, many human-origin H3N2 viruses were also found between the middle of the 1970s and the middle of the 1980s (Yu et al., 2008). A number of H1N2 viruses, both short-lived and long-lived, have also been discovered, although they are generally less prevalent than other subtypes (Cheung et al., 2023). One particularly unusual variation is the H1N7 virus, which appears to be a hybrid of horse and SIVs (Brown et al., 1994). Currently, there are no viruses in Europe that carry the triple-reassortant internal gene (TRIG), and the diversity of these viruses is believed to be lower than that in North America (Lorusso et al., 2013). Only 3% of the viruses discovered from pigs during surveillance in a number of countries between 2006 and 2008 were novel viruses (Abe et al., 2015). Different SIV strains may be more prevalent in different places, and some viruses can only temporarily infect Asian pigs. A significant outbreak in Japan in 1989–1990 was caused by an H1N2 virus that originated in Asia, propagated among Japanese pigs, and has since spread to a number of other nations (Ouchi et al., 1996). Information on SIV in Africa, Central and South America, and Mexico is currently lacking. Although H3N2 and H1N1 viruses are known to spread throughout Latin America, little is known about their genetic makeup (Komadina et al., 2014). Although the H3N2 virus is highly contagious in pigs, one isolate from an Argentine respiratory disease outbreak was completely derived from a human influenza virus (Cappuccio et al., 2011). One report from Africa mentions H1 viruses, and a recent study from Cameroon discovered the 2009 pandemic H1N1 virus in pigs grown in the wild (Njabo et al., 2012). This could potentially occur in other continents and regions. In the United States, swine influenza is a prevalent concern, with studies indicating an individual pig prevalence of 4.6% and a herd prevalence of 90.6% over a 12–24 month period. The primary circulating subtypes are H1N1, H1N2, and H3N2. Notably, in October 2024, the USDA National Veterinary Services Laboratories confirmed the first detection of H5N1 bird influenza in a pig on a backyard farm in Oregon. While the risk to the public was deemed low, this incident underscored the potential for interspecies transmission and the necessity for vigilant monitoring (Li and Robertson, 2021). European swine populations are primarily affected by H1N1, H1N2, and H3N2 subtypes. A comprehensive study across Belgium, France, Italy, and Spain reported a 90% farm-level prevalence, with individual pig seroprevalence at 62%. The epidemiology is complex and is influenced by factors such as the cocirculation of viruses, prior immunity, and husbandry practices. Surveillance programs have revealed significant genetic diversity among these viruses, contributing to evolving epidemiological patterns (Brown, 2013). In China and Southeast Asia, swine influenza is enzootic, with multiple subtypes, including H1, H3, H4, H5, H7, and H9, circulating within pig populations (Li and Robertson, 2021). Data on swine influenza in Africa is limited. However, the continent has faced significant challenges from other swine diseases, notably African Swine Fever. Host rangeHumans, pigs, horses, marine animals, and birds are among the many animal species that are infected by IAVs (Taubenberger and Kash, 2010). There is a significant chance that influenza viruses may spread from one species to another in nature due to global interactions among people, pigs, birds, and other mammalian species (Short et al., 2015). Although pigs are the primary hosts of SIV, some viruses can also infect turkeys, ferrets, and minks (Abdelwhab and Mettenleiter, 2023). A duck in Hong Kong was found to have an H1N1 SIV, which is virulent to pigs and poultry and can infect ducks in an experiment (Suarez et al., 1998). Calves are also known to have this virus (Lopez and Woods, 1987). Although the source of the virus could not be definitively identified, a subsequent investigation indicated that cattle’s antibodies to H3 viruses were probably brought on by exposure to SIV (Sreenivasan et al., 2019). Pigs are special hosts for both human and bird species; they are mixed hosts that produce strains adapted for humans (Brown, 2001). Pathogenesis and pathologyThe attachment and reproduction of SIVs are often limited to the pig respiratory system. The process of SIV infection is similar to that of respiratory viral infections: the virus enters the body via the aerosol, and the infection spreads quickly to the nasal cavity and epithelium of major airways (Zhang et al., 2013). In a few hours, the virus may spread to every airway. When the virus is inhaled, it sticks to the lower respiratory tract’s surface and causes exudation, focal atelectasis, focal necrosis of the bronchial epithelium, and severe lung hyperemia (Khatri et al., 2010). The lung regions afflicted by the pneumonic lesions were purple, rigid, and swollen with interlobular edema, according to the postmortem examination (Cunha, 2010). There is swelling and edema of the lymph nodes (bronchial, cervical, mediastinal, and mesenteric) may occur, and the airways may be filled with blood-tinged fibrinous exudate (Weingartl et al., 2009). In severe cases, fibrinous pleurisy and interstitial pneumonia develop (Cunha, 2010). Microscopic lesions include isolated atelectasis and emphysema, thickened alveolar septa, exudative bronchitis, interstitial pneumonia, peribronchial and perivascular cellular infiltration, pulmonary parenchymal congestion, and degenerative and necrotic alterations in the bronchial epithelium (Sarli et al., 2021). In a recent investigation on pathogenicity in particular pathogen-free pigs, it was found that H1N1-infected pigs had higher lung lesion scores than H3N2-infected pigs (Sreta et al., 2009). However, both groups had lesions that included peribronchial and perivascular mononuclear cell infiltration, airway blockage, and epithelial cell injury. Immune responseImmune responses such as innate, mucosal, and systemic immunity (both humoral and cellular immunity) are triggered by IAV infection. Early influenza virus infection containment depends on innate immunity, the host’s initial line of defense that nonspecifically prevents influenza virus infection (Chen et al., 2018). Numerous innate inhibitors soluble in respiratory secretions are part of the complex innate immune response, which plays a major role in directing and promoting adaptive and pathogen-specific immune responses (White et al., 2008). The influenza virus first adheres to the respiratory tract’s mucosal tissue to start an illness. The type I IFN system is triggered to create an antiviral state within the cell when Toll-like receptors or cytoplasmic sensors (such as retinoic acid-inducible gene I and melanoma differentiation-associated gene 5) recognize viral products such as viral RNA within the cell (Baum and García-Sastre, 2010). In addition to innate immunity, the mucosal immune response offers a crucial line of defense against influenza infection in animals that have previously been exposed to or vaccinated against influenza viruses (Mettelman et al., 2022). The primary neutralizing antibodies that stop influenza virus entry and can stop intracellular influenza replication are specific IgA and IgM, which are locally released in the respiratory tract (van de Sandt et al., 2012). The HA and NA surface proteins of influenza viruses are the specific targets of neutralizing antibodies found in nasal secretions (Zhang et al., 2019). Mucosal antibodies specific to influenza play a major role in the removal of SIV from the respiratory system in a pig model (Richt et al., 2006). In the respiratory tract, mucosal immunity induced by natural influenza infection is more effective and protective against subsequent heterovariant virus infection than systemic immunity induced by parenteral immunization with inactivated vaccines. The humoral immune system generates antibodies against all of the main influenza virus proteins while an infection is present (Bahadoran et al., 2016). Antibodies against HA are crucial for neutralizing viruses and preventing illness (Ohshima et al., 2011). The discharge of mature viruses from infected cells is inhibited by antibodies against NA, but they are less successful in stopping infection (Pantaleo et al., 2022). The M2 protein may play a part in antibody-mediated protection, although antibodies against conserved internal proteins (M and NP) may not offer defense against infection (Staneková and Varečková, 2010). Serum HA and NA antibody levels are believed to correlate with disease prevention and resistance because these antibodies are the most crucial for protection against influenza (Desheva et al., 2024). However, if the antigenic shift or the infecting virus changes, the humoral immune response may not be able to stop influenza infection. The removal of influenza viruses from the respiratory system and subsequent recovery from the illness are significantly influenced by cell-mediated immunity (Denney and Ho, 2018). The blood and lower respiratory tract of infected hosts contain influenza-specific cytotoxic T lymphocytes (CTLs), which can lyse cells infected with different IAV subtypes (Chen et al., 2018). Certain CTL responses in people and animals target internal influenza viral proteins, specifically NP, M1, NS1, and polymerase proteins (PB1, PB2, and PA) (Jameson et al., 1998). In mice and humans, IAV viral nucleoprotein (NP) is a crucial target antigen for CTLs that are cross-reactive and subtype-specific (Rak et al., 2023). Currently, little information is available on how pig cellular immune systems react to influenza infection. Prior research has demonstrated that CTL responses, when combined with neutralizing antibodies, are essential for lowering virus shedding and eliminating virus because they are cross-reactive among IAV strains, providing heterovariant and heterosubtype immunity (Grebe et al., 2008). Consequently, a perfect vaccination can elicit a well-rounded immune response that includes humoral, cellular, and mucosal immunity. Clinical signsIn pigs: Clinical signs appear rapidly. Infected pigs exhibit symptoms like coughing, sneezing, nasal discharge, tachypnea, dyspnea, fever, anorexia, and lethargy (Sreta et al., 2009). Some may also occasionally exhibit signs of deadly bronchopneumonia, and pregnant sows may abort (Galwankar and Clem, 2009). Clinical symptoms typically last for seven days, after which there is a quick and full recovery, unless there is a subsequent bacterial infection that exacerbates the symptoms. This disease causes high morbidity (up to 100%) but low mortality if secondary bacterial infections are avoided (Cunha, 2010). In herds in which infection is a persistent issue or all animals have received vaccination, there may be a chance of sporadic disease with few or no symptoms. In humans: Typical signs appear after an incubation period of 1–7 days, most often affecting adolescents. People who contract new swine influenza infections have bexperienceiety of symptoms that resemble those of seasonal influenza (Peiris et al., 2007). The primary symptoms are fever, chills, headache, runny nose, body aches, joint pain or myalgia, cough, sore throat, and exhaustion (Hasan et al., 2018). Additionally, diarrhea, vomiting, and nausea can occur. Like seasonal flu, major flu-related complications can be noticed in the elderly, children, pregnant women, and persons with asthma, diabetes, heart disease, or immunosuppression (Lim and Mahmood, 2011). However, those aged 5–25 years accounted for the majority of infections during the current swine influenza outbreak (Altayep et al., 2017). The signs of flu in children, such as shortness of breath, bluish or gray skin, severe or prolonged vomiting, irritability, lack of interaction, and other general flu-like symptoms (Rewar et al., 2015). Important symptoms in adults include shortness of breath or difficulty breathing, chest or stomach pain, abrupt lightheadedness, and acute or ongoing vomiting (Al Hajjar and McIntosh, 2010). DiagnosisReverse transcription polymerase chain reaction (RT-PCR) detection technology can be used to quantify mRNA and determine an individual’s viral load (Ho-Pun-Cheung et al., 2009). The HA sequence of SIVs is the primary basis for the development of PCR tests. This test can extract RNA from influenza viruses by amplifying and detecting nasopharyngeal samples (Schorr et al., 1994). Moreover, throat swabs and bronchial aspirates can be used as samples. To identify IAV infection, respiratory specimens from infected individuals must be obtained 4–5 days after the onset of symptoms (Eisfeld et al., 2014). Swine H1N1 viruses can be distinguished from other seasonal viral illnesses thanks to the test procedure’s high specificity (Ravina et al., 2020). Results from this test can be obtained within a few hours of the sample being collected. The virus is produced on a culture medium in a virus culture technique, which allows for a 100% precise diagnosis of SIV (Detmer et al., 2013). The proposed method can obtain a negative predictive value of approximately 90% and high sensitivity. Within 2–3 days of inoculation, the viral strain is cultured in chicken embryo and monkey kidney cell cultures (Zhang and Gauger, 2014). Rapid diagnostic techniques require respiratory secretions with high virus concentrations, yet they have a sensitivity of 60%–80% and can identify viral antigens by comparing them with regular RT-PCR data collected from 65 individuals (Vasoo et al., 2009). A clinical suspicion of the disease was used to interpret a negative report. The Antibody Titer Rise Test is mostly used to examine past cases; however, it is not used for diagnostic purposes (Detmer et al., 2013). This method also aids in diagnosis by comparing antibody titer levels obtained during the acute phase of the illness with those obtained 10–14 days later. Hematological and biological tests may reveal elevated levels of creatine kinase, leukopenia, and lactate dehydrogenase (Wang et al., 2014). In addition, uncommon signs of thrombocytopenia. Abnormalities on chest X-rays can be noticed, particularly in hospitalized and severely affected individuals (Agarwal et al., 2009). TransmissionIn mammals, influenza viruses are spread by direct or indirect contact with nasal secretions, as well as by droplets and aerosols released during coughing and sneezing (Richard and Fouchier, 2016). Direct contact between animals that are infected and those that are not is the primary method of transmission (van Diemen et al., 2023). Due to the close quarters in which pigs are housed, the virus can spread directly between pigs via contact between their noses or through dried mucus (Galwankar and Clem, 2009). Another significant form of infection is airborne transmission via aerosols released by pigs that cough or sneeze (Hu et al., 2023). Figure 1 depicts the transmission, including direct contact, aerosol spread, and via fomites. Pigs are commonly implicated in the transmission of influenza viruses between species and are the main reservoirs of H1N1 and H3N2 influenza viruses (Brown, 2001). The development of human influenza pandemic strains may be influenced by the presence of viruses in pigs and the regular introduction of novel viruses from other animals (Anderson et al., 2021). Laboratory staff were then exposed to the swine H1N1 virus isolated from Turkey (Hinshaw et al., 1983). According to these results, viruses from people, pigs, ducks, and turkeys can potentially spread to one another. Economic losses resulted from the spread of the avian H1N1 virus to pigs that were already on the market in Europe and then returned to Turkey from the pigs (Ludwig et al., 1994). Human-to-pig transmission occurs (Ma, 2020), regardless of where the virus originated; once a herd is infected, it continues to spread through the introduction of new herds and the birth of susceptible young pigs, frequently leading to endemically infected herds (Lagan et al., 2024). Pigs may potentially develop new virus lineages as a result of the transmission of viruses from other animals being transmitted to them (Everett et al., 2020). Figure 1 also highlights zoonotic transmission pathways from pigs to humans and other animals, emphasizing the role of pigs in swine influenza. Risk factorsSwine influenza most commonly attacks children aged 5 years and over and teenagers (O’Riordan et al., 2010). This is a remarkable finding because older adults persons and the very young are more likely to experience problems from most influenza virus infections. The risk factors of catching swine influenza are currently the same as those of other flus. Spending time in places where a large number of people have swine influenza puts them at the most risk (Mastin et al., 2011). Infection with swine influenza puts some persons at greater risk of developing a serious illness. These groups include pregnant women, children under 5 years old, adolescents and children under 19 years old receiving long-term aspirin therapy, adults over 65 years old, people with weakened immune systems (from conditions like AIDS), and people with chronic conditions like diabetes mellitus, asthma, heart disease, or neuromuscular disease (del Rio and Guarner, 2010).
Fig. 1. The transmission pathways of swine influenza. Swine influenza spreads among pigs through direct contact, respiratory droplets, fomites, and vertical transmission. Zoonotic transmission involves close contact with aerosols, aerosols, and contaminated materials. Swine influenza is also transmitted to other mammals. Public health importanceBoth people and animals have experienced considerable illness and mortality as a result of emerging zoonotic diseases. Swine influenza is a worldwide viral zoonotic illness that spreads quickly (Vincent et al., 2008). There are reports of this illness from numerous nations worldwide. Infected pigs can spread infection to humans through close contact (Dandagi and Byahatti, 2011). This illness is contracted by coughing, sneezing, contact, and occasionally contaminated things (Cunha, 2010). Pigs can contract this disease from sick people. SIVs are common in pig populations worldwide (Mastin et al., 2011). Pig-to-human viral transmission is rare and frequently results in the generation of antibodies in the blood, rather than causing influenza in people (Mo et al., 2022). The infection is referred to as zoonotic swine influenza if it results in human influenza. Infections with human SIV have been documented in Europe (Fraaij et al., 2016), Asia (Adisasmito et al., 2019), Canada (Forgie et al., 2011), and the United States (Olsen et al., 2002). The human swine influenza and common influenza are not distinguished by any particular clinical characteristics. Healthy persons are also at a high risk of contracting swine influenza and dying from it, even if some case patients have immune-compromising predisposing factors (Guillari et al., 2021). Strong case ascertainment bias is probably the cause of the high percentage of deaths in this case series. In line with sero-epidemiological studies that have demonstrated elevated rates of SIV infection in individuals with occupational exposure to pigs, the majority of case patients reported contact with pigs (Rovida et al., 2016). Additionally, individuals who work with pigs may serve as a conduit for the virus’s spread throughout their communities. The discovery of influenza subtypes around the middle of the 20th century made it feasible to accurately diagnose human transmission. There have only been 50 verified cases of these diseases since then (Kilbourne, 2006). Human-to-human transmission of the strain of swine influenza is uncommon. The symptoms of influenza and influenza-like illnesses in general are comparable to those of zoonotic swine influenza in humans (Hennig et al., 2022). Human infections with several SIVs H1N1, H3N2, and H1N2 have been documented occasionally (Robinson et al., 2007; Schnitzler and Schnitzler, 2009; Bravo-Vasquez et al., 2017). Certain genotypes may have a higher propensity to infect people. There is evidence that certain SIVs can spread from person to person, including a significant outbreak at the Fort Dix military base in the 1970s (Lessler et al., 2007). It is the only virus from the 2009 H1N1 pandemic to have adapted to humans. Economic impactThe 2009 H1N1 outbreak, also known as the swine influenza, had a significant impact on the global economy. Pork markets both domestically and internationally declined as a result of the outbreak (McLean and Graham, 2022). As a result of consumers’ fear of eating pork, demand and prices have fallen precipitously. According to estimates, the 2009 H1N1 virus decreased the world’s gross domestic product by 0.5% to 1.5% (Smith et al., 2009). The epidemic has increased direct expenses for care, such as prescription drugs, hospital stays, and outpatient visits. For instance, hospitalization expenses for every patient with H1N1 in Canada were approximately $11,000 (Ng et al., 2018). The epidemic has resulted in significant economic hardship and social unrest. The economy may also be impacted by factors like sick workers who are less productive and preventative actions like preventing individuals from going to work to prevent infections that affect the labor supply (Borkenhagen et al., 2020). TreatmentPigs seldom die from swine influenza; therefore, supportive care and relaxation are the only treatments required (Ma, 2020). Rather, the goal of veterinary care is to stop the infection from spreading to adjacent farms or throughout the farm. Techniques for animal management and vaccination are essential to this endeavor (Salvesen and Whitelaw, 2021). This illness is treated with antibiotics, which help prevent bacterial pneumonia and other secondary illnesses in calves weakened by influenza, even if they have little impact on the influenza virus (Moghadami, 2017). Similar to human influenza, SIV-induced illness is treated with supportive treatment (such as rest and fluids) in less complicated cases, antibiotics when necessary for subsequent bacterial pneumonia, and hospitalization if necessary for more severe cases (Rewar et al., 2015). Two classes of antiviral medications can block IAV: NA inhibitors (zanamivir, oseltamivir, peramivir, and laninamivir) and adamantane (amantadine, rimantadine) (Hussain et al., 2017). The use of these medications within the first 48 hours of the beginning of clinical symptoms maximizes their effectiveness. Although the effectiveness of amantadine against SIV is unknown, amantadine-resistant isolates are prevalent in virus lineages found in US pigs (Smyk et al., 2022). Treatment of the disease includes bed rest, nutrition (enteral or parenteral as tolerated), broad-spectrum antibiotics to treat or prevent secondary bacterial pneumonia (mostly Gram-positive), electrolyte balance, oral fluids (intravenous in severe cases), oxygen therapy or in severe persistent hypoxia the use of ventilator support or extracorporeal oxygenation in cases of severe refractory acute respiratory distress syndrome, and vasopressors for shock (Rewar et al., 2015). Aspirin and other salicylates are prohibited during influenza because they can cause Reye’s syndrome (Noor and Gradidge, 2018). It has been demonstrated that using large dosages of corticosteroids is not beneficial and may even be harmful (low doses of 200 mg/day may be utilized in cases of refractory septic shock) (Zhang et al., 2015). Oseltamivir has been shown in numerous studies to be effective against seasonal influenza in both pre- and post-exposure prophylaxis, with 68%–89% efficacy when administered to household contacts (Dutkowski, 2010; Ison, 2013). Nonetheless, it demonstrated 74% efficacy in 6 weeks of pre-exposure prophylaxis. Consequently, all patients exposed to influenza should receive postexposure chemoprophylaxis. Additionally, because health care workers are likely to come into touch with swine influenza cases, they are strongly encouraged to obtain chemoprophylaxis (Galwankar and Clem, 2009). VaccinationThe SIV vaccines now in use might not be able to fully eradicate the clinical symptoms of infection in pigs or produce robust immunity. The H1N1, H3N2, and H1N2 subtypes, which have antigenically distinct HAs, are already circulating in pigs worldwide, making a vaccination that can produce cross-protective immunity between various subtypes and strains imperative (Takemae et al., 2013). However, relying solely on vaccination methods to prevent swine influenza may not be the best option due to the growing number of novel subtypes and genetic variations. Although pig vaccination is not always successful, it can lower the rate at which infected animals shed the virus, thus lowering the risk of zoonotic infection and human exposure (Monath, 2013). Pigs can be protected against either the recombinant equine herpesvirus (EHV-1) or other influenza viruses by expressing H1 from A(H1N1)pdm09 (Said et al., 2013). A vaccination against the current strain of the H1N1 pandemic has recently been produced and approved for use in the UK by the Food and Drug Administration and the European Medicines Agency (Nuwarda et al., 2021). These include cell culture vaccines (Vero or MDCK), influenza viruses derived from influenza viruses, and SIV cultivated from chicken eggs, which are used as lethal vaccines or subunit vaccinations following digestion with detergent (Hegde, 2015). Human influenza vaccinations do not protect recipients from avian influenza, and they may cause adverse effects following classical influenza vaccinations. There are two kinds of influenza vaccines: the nasal spray-based Live Attenuated Influenza Vaccine (LAIV) and the injection-based Trivalent Inactivated Influenza Vaccine. People under the age of two and those over the age of 49 are not advised to use LAIV (Hoft et al., 2011). There is currently a need for efficient vaccination to shield humans and birds from the novel H1N1 and H5N1 virus subtypes (Dey et al., 2023). The WHO advises that any medical personnel who have come into contact with suspected or confirmed swine flu infections should be vaccinated (doctors, nurses, paramedics, and ambulance staff) (Galwankar and Clem, 2009). There is now an injectable vaccine for influenza A. It takes around two to three weeks for the body’s immune response to develop following immunization; in the interim, preventive chemotherapy might be employed (Petro-Turnquist et al., 2024). ControlVeterinarians should be aware of local reporting regulations even though SIVs are ubiquitous and widespread among pigs and typically do not necessitate reporting (Olsen et al., 2002). Consultation with state authorities is required. Total manufacturing is a management strategy that can help prevent viruses from entering the system. Newly purchased pigs or animals brought back to the facility must also be isolated and tested (Alarcón et al., 2021). A biosecurity plan should also account for contact with wild and feral pigs, wild birds (particularly waterfowl and other aquatic birds), poultry, humans, and potentially other species like horses. In addition, fomites include contaminated water sources that may harbor viruses (Foni et al., 2013). In herds that are consistently afflicted, good management can help lessen the severity of the disease, and early isolation of sick animals can help lessen transmission inside the facility (Mastin et al., 2011). Depopulation can eradicate the influenza virus from infected pig herds (Er et al., 2016). Some managerial initiatives may also be effective. Hand hygiene and sanitation (e.g., frequent hand washing) and the use of personal protective equipment, where suitable, are preventative measures against zoonotic influenza viruses (Moncion et al., 2019). Numerous public health organizations have released comprehensive guidelines, which include advice for the general public and agricultural fair exhibitors. These guidelines, in general, emphasize handwashing, avoiding close contact with pigs, and taking preventative measures to prevent mucous membrane contamination, like forbidding eating and drinking in pig pens (Dandagi and Byahatti, 2011). Additionally, they urge small children and anyone who is susceptible to more severe sickness from human seasonal influenza viruses to avoid pigs and pig enclosures at fairs. Although it is unlikely that retail meat will contain live SIVs, heating the meat will inactivate any viruses that make it to the consumer. In addition, viruses on fomites can be eliminated by following standard food safety procedures when handling raw meat products (Shurson et al., 2023). The possibility of zoonotic exposure should be taken into account when seeing a doctor for a disease that develops shortly after coming into contact with an animal. ConclusionSwine influenza, a respiratory disease caused by the SIV, poses significant health risks to both pigs and humans, leading to severe outbreaks and financial losses in the agricultural sector. Understanding the virus’s transmission dynamics and developing effective management strategies are crucial for mitigating its impact on public health and livestock production. AcknowledgmentsThe authors would like to thank Universitas Airlangga and Badan Riset dan Inovasi Nasional. Author’s contributionsDKM, ARK, SW, MKJK, and BWKW drafted the manuscript. LA, RZA, IBM, and AOA revised and edited the manuscript. SU, IF, WW, and DAAK participated in preparing and critically checking this manuscript. RR, TDL, IM, FE, and SM edited the references. All authors have read and approved the final manuscript. Conflict of interestThe authors declare no conflict of interest. FundingThe authors thank Universitas Airlangga for managerial support, Salma Firdausya Qurrotunnada Noor, Eunice Wong Hui Wen, Joo Jia Yin, and Rahma Novhira for technical support. This research was funded by the Directorate of Research and Community Service, Deputy for Strengthening Research and Technology, Ministry of Research and Technology/National Research and Innovation Agency for the 2022 fiscal year, Chancellor’s Decree number: 770/UN3.14/PT/2022. Data availabilityAll references are open-access, so data can be obtained from the online web. ReferencesAbdelwhab, E.M. and Mettenleiter, T.C. 2023. Zoonotic animal influenza virus and potential mixing vessel hosts. Viruses 15(4), 980. Abe, H., Mine, J., Parchariyanon, S., Takemae, N., Boonpornprasert, P., Ubonyaem, N., Patcharasinghawut, P., Nuansrichay, B., Tanikawa, T., Tsunekuni, R. and Saito, T. 2015. Co-infection of influenza A viruses of swine contributes to effective shuffling of gene segments in a naturally reared pig. Virology 484(1), 203–212. Adisasmito, W., Budayanti, S., Aisyah, D.N., Coker, R., Andayani, A.R., Smith, G.J.D. and Rudge, J.W. 2019. Surveillance and characterisation of influenza viruses among patients with influenza-like illness in Bali, Indonesia, July 2010-June 2014. BMC Infect. Dis. 19(1), 231. Agarwal, P.P., Cinti, S. and Kazerooni, E.A. 2009. Chest radiographic and CT findings in novel swine-origin influenza A (H1N1) virus (S-OIV) infection. AJR Am. J. Roentgenol. 193(6), 1488–1493. Al Hajjar, S. and McIntosh, K. 2010. The first influenza pandemic of the 21st century. Ann. Saudi Med. 30(1), 1–10. Alarcón, L.V., Allepuz, A. and Mateu, E. 2021. Biosecurity in pig farms: a review. Porc. Health Manag. 7(1), 5. Allen, J.D. and Ross, T.M. 2018. H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum. Vaccin. Immunother. 14(8), 1840–1847. Altayep, K.M., Ahmed, H.G., A Tallaa, A.T., Alzayed, A.S., Alshammari, A.J. and Talla, A.T.A. 2017. Epidemiology and clinical complication patterns of influenza A (H1N1 Virus) in Northern Saudi Arabia. Infect. Dis. Rep. 9(2), 6930. Anderson, T.K., Chang, J., Arendsee, Z.W., Venkatesh, D., Souza, C.K., Kimble, J.B., Lewis, N.S., Davis, C.T. and Vincent, A.L. 2021. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb. Perspect. Med. 11(3), a038737. Anhlan, D., Grundmann, N., Makalowski, W., Ludwig, S. and Scholtissek, C. 2011. Origin of the 1918 pandemic H1N1 influenza A virus as studied by codon usage patterns and phylogenetic analysis. RNA 17(1), 64–73. Anjorin, A.A., Sausy, A., Muller, C.P., Hübschen, J.M., Omilabu, S.A. and Snoeck, C.J. 2023. Human seasonal influenza viruses in swine workers in Lagos, Nigeria: consequences for animal and public health. Viruses 15(6), 1219. Bahadoran, A., Lee, S.H., Wang, S.M., Manikam, R., Rajarajeswaran, J., Raju, C.S. and Sekaran, S.D. 2016. Immune responses to influenza virus and its correlation to age and inherited factors. Front. Microbiol. 7(1), 1841. Baum, A. and García-Sastre, A. 2010. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids 38(5), 1283–1299. Bhatta, T.R., Ryt-Hansen, P., Nielsen, J.P., Larsen, L.E., Larsen, I., Chamings, A., Goecke, N.B. and Alexandersen, S. 2020. Infection dynamics of swine influenza virus in a danish pig herd reveals recurrent infections with different variants of the H1N2 swine influenza a virus subtype. Viruses 12(9), 1013. Borkenhagen, L.K., Wang, G.L., Simmons, R.A., Bi, Z.Q., Lu, B., Wang, X.J., Wang, C.X., Chen, S.H., Song, S.X., Li, M., Zhao, T., Wu, M.N., Park, L.P., Cao, W.C., Ma, M.J. and Gray, G.C. 2020. High risk of influenza virus infection among swine workers: examining a dynamic cohort in China. Clin. Infect. Dis. 71(3), 622–629. Bouvier, N.M. and Palese, P. 2008. The biology of influenza viruses. Vaccine 26 (Suppl 4), D49–D53. Bravo-Vasquez, N., Karlsson, E.A., Jimenez-Bluhm, P., Meliopoulos, V., Kaplan, B., Marvin, S., Cortez, V., Freiden, P., Beck, M.A., Hamilton-West, C. and Schultz-Cherry, S. 2017. Swine influenza virus (H1N2) characterization and transmission in Ferrets, Chile. Emerg. Infect. Dis. 23(2), 241–251. Brockwell-Staats, C., Webster, R.G. and Webby, R.J. 2009. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza Other Respir. Viruses 3(5), 207–213. Brown, I.H. 2001. The pig as an intermediate host for influenza A viruses between birds and humans. Int. Congr. Ser. 1219(1), 173–178. Brown, I.H. 2013. History and epidemiology of Swine influenza in Europe. Curr. Top. Microbiol. Immunol. 370(1), 133–146. Brown, I.H., Alexander, D.J., Chakraverty, P., Harris, P.A. and Manvell, R.J. 1994. Isolation of an influenza A virus of unusual subtype (H1N7) from pigs in England, and the subsequent experimental transmission from pig to pig. Vet. Microbiol. 39(1–2), 125–134. Brown, I.H., Harris, P.A. and Alexander, D.J. 1995. Serological studies of influenza viruses in pigs in Great Britain 1991-2. Epidemiol. Infect. 114(3), 511–520. Cappuccio, J.A., Pena, L., Dibárbora, M., Rimondi, A., Piñeyro, P., Insarralde, L., Quiroga, M.A., Machuca, M., Craig, M.I., Olivera, V., Chockalingam, A., Perfumo, C.J., Perez, D.R. and Pereda, A. 2011. Outbreak of swine influenza in Argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J. Gen. Virol. 92(Pt 12), 2871–2878. Chen, X., Liu, S., Goraya, M.U., Maarouf, M., Huang, S. and Chen, J.L. 2018. Host immune response to influenza A virus infection. Front. Immunol. 9(1), 320. Cheung, J., Bui, A.N., Younas, S., Edwards, K.M., Nguyen, H.Q., Pham, N.T., Bui, V.N., Peiris, M. and Dhanasekaran, V. 2023. Long-term epidemiology and evolution of swine Influenza viruses, Vietnam. Emerg. Infect. Dis. 29(7), 1397–1406. Cunha, B.A. 2010. Swine Influenza (H1N1) pneumonia: clinical considerations. Infect. Dis. Clin. North Am. 24(1), 203–228. Dandagi, G.L. and Byahatti, S.M. 2011. An insight into the swine-influenza A (H1N1) virus infection in humans. Lung India 28(1), 34–38. del Rio, C. and Guarner, J. 2010. The 2009 influenza A (H1N1) pandemic: what have we learned in the past 6 months. Trans. Am. Clin. Climatol. Assoc. 121(1), 128–137 Denney, L. and Ho, L.P. 2018. The role of respiratory epithelium in host defence against influenza virus infection. Biomed. J. 41(4), 218–233. Desheva, Y., Sergeeva, M., Kudar, P., Rekstin, A., Romanovskaya-Romanko, E., Krivitskaya, V., Kudria, K., Bazhenova, E., Stepanova, E., Krylova, E., Kurpiaeva, M., Lioznov, D., Stukova, M. and Kiseleva, I. 2024. Neuraminidase antibody response to homologous and drifted influenza A viruses after immunization with seasonal influenza vaccines. Vaccines (Basel) 12(12), 1334. Detmer, S., Gramer, M., Goyal, S., Torremorell, M. and Torrison, J. 2013. Diagnostics and surveillance for Swine influenza. Curr. Top. Microbiol. Immunol. 370(1), 85–112. Dey, P., Ahuja, A., Panwar, J., Choudhary, P., Rani, S., Kaur, M., Sharma, A., Kaur, J., Yadav, A.K., Sood, V., Babu, A.R.S., Bhadada, S.K., Singh, G., Barnwal, R.P. 2023. Immune control of avian influenza virus infection and its vaccine development. Vaccines (Basel) 11(3), 593. Dou, D., Revol, R., Östbye, H., Wang, H. and Daniels, R. 2018. Influenza A virus cell entry, replication, virion assembly and movement. Front. Immunol. 9(1), 1581. Dutkowski, R. 2010. Oseltamivir in seasonal influenza: cumulative experience in low- and high-risk patients. J. Antimicrob. Chemother. 65(Suppl 2), ii11–ii24. Eisfeld, A.J., Neumann, G. and Kawaoka, Y. 2014. Influenza A virus isolation, culture and identification. Nat. Protoc. 9(11), 2663–2681. Er, C., Skjerve, E., Brun, E., Framstad, T. and Lium, B. 2016. Occurrence and spread of influenza A(H1N1)pdm09 virus infection in Norwegian pig herds based on active serosurveillance from 2010 to 2014. Epidemiol. Infect. 144(15), 3148–3165. Everett, H.E., Nash, B., Londt, B.Z., Kelly, M.D., Coward, V., Nunez, A., van Diemen, P.M., Brown, I.H. and Brookes, S.M. 2020. Interspecies transmission of reassortant swine influenza A virus containing genes from swine influenza A(H1N1)pdm09 and A(H1N2) viruses. Emerg. Infect. Dis. 26(2), 273–281. Foni, E., Garbarino, C., Chiapponi, C., Baioni, L., Zanni, I. and Cordioli, P. 2013. Epidemiological survey of swine influenza A virus in the wild boar population of two Italian provinces. Influenza Other Respir. Viruses 7(Suppl 4), 16–20. Forgie, S.E., Keenliside, J., Wilkinson, C., Webby, R., Lu, P., Sorensen, O., Fonseca, K., Barman, S., Rubrum, A., Stigger, E., Marrie, T.J., Marshall, F., Spady, D.W., Hu, J., Loeb, M., Russell, M.L. and Babiuk, L.A. 2011. Swine outbreak of pandemic influenza A virus on a Canadian research farm supports human-to-swine transmission. Clin. Infect. Dis. 52(1), 10–18. Fraaij, P.L., Wildschut, E.D., Houmes, R.J., Swaan, C.M., Hoebe, C.J., de Jonge, H.C., Tolsma, P., de Kleer, I., Pas, S.D., Munnink, B.B.O., Phan, M.V., Bestebroer, T.M., Roosenhoff, R.S., van Kampen, J.J., Cotton, M., Beerens, N., Fouchier, R.A., van den Kerkhof, J.H., Timen, A. and Koopmans, M.P. 2016. Severe acute respiratory infection caused by swine influenza virus in a child necessitating extracorporeal membrane oxygenation (ECMO), the Netherlands, October 2016. Euro Surveill. 21(48), 30416. Galwankar, S. and Clem, A. 2009. Swine influenza A (H1N1) strikes a potential for global disaster. J. Emerg. Trauma Shock 2(2), 99–105. Gray, G.C., McCarthy, T., Capuano, A.W., Setterquist, S.F., Olsen, C.W. and Alavanja, M.C. 2007. Swine workers and swine influenza virus infections. Emerg. Infect. Dis. 13(12), 1871–1878. Grebe, K.M., Yewdell, J.W. and Bennink, J.R. 2008. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect. 10(9), 1024–1029. Guillari, A., Polito, F., Pucciarelli, G., Serra, N., Gargiulo, G., Esposito, M.R., Botti, S., Rea, T. and Simeone, S. 2021. Influenza vaccination and healthcare workers: barriers and predisposing factors. Acta Biomed. 92(S2), e2021004. Hasan, F., Khan, M.O. and Ali, M. 2018. Swine flu: knowledge, attitude, and practices survey of medical and dental students of Karachi. Cureus 10(1), e2048. Hegde, N.R. 2015. Cell culture-based influenza vaccines: a necessary and indispensable investment for the future. Hum. Vaccin. Immunother. 11(5), 1223–1234. Hennig, C., Graaf, A., Petric, P.P., Graf, L., Schwemmle, M., Beer, M. and Harder, T. 2022. Are pigs overestimated as a source of zoonotic influenza viruses? Porcine Health Manag. 8(1), 30. Henritzi, D., Petric, P.P., Lewis, N.S., Graaf, A., Pessia, A., Starick, E., Breithaupt, A., Strebelow, G., Luttermann, C., Parker, L.M.K., Schröder, C., Hammerschmidt, B., Herrler, G., Beilage, E.G., Stadlbauer, D., Simon, V., Krammer, F., Wacheck, S., Pesch, S., Schwemmle, M., Beer, M. and Harder, T.C. 2020. Surveillance of European domestic pig populations identifies an emerging reservoir of potentially zoonotic swine influenza A viruses. Cell Host Microbe 28(4), 614–627.e6. Herfst, S., Zhang, J., Richard, M., McBride, R., Lexmond, P., Bestebroer, T.M., Spronken, M.I.J., de Meulder, D., van den Brand, J.M., Rosu, M.E., Martin, S.R., Gamblin, S.J., Xiong, X., Peng, W., Bodewes, R., van der Vries, E., Osterhaus, A.D.M.E., Paulson, J.C., Skehel, J.J. and Fouchier, R.A.M. 2020. Hemagglutinin traits determine transmission of avian A/H10N7 influenza virus between mammals. Cell Host Microbe 28(4), 602–613.e7. Hinshaw, V.S., Webster, R.G., Bean, W.J., Downie, J. and Senne, D.A. 1983. Swine influenza-like viruses in turkeys: potential source of virus for humans? Science 220(4593), 206–208. Hoft, D.F., Babusis, E., Worku, S., Spencer, C.T., Lottenbach, K., Truscott, S.M., Abate, G., Sakala, I.G., Edwards, K.M., Creech, C.B., Gerber, M.A., Bernstein, D.I., Newman, F., Graham, I., Anderson, E.L. and Belshe, R.B. 2011. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J. Infect. Dis. 204(6), 845–853. Ho-Pun-Cheung, A., Bascoul-Mollevi, C., Assenat, E., Boissière-Michot, F., Bibeau, F., Cellier, D., Ychou, M. and Lopez-Crapez, E. 2009. Reverse transcription-quantitative polymerase chain reaction: description of a RIN-based algorithm for accurate data normalization. BMC Mol. Biol. 10(1), 31. Horimoto, T. and Kawaoka, Y. 2001. Pandemic threat posed by avian influenza A viruses. Clin. Microbiol. Rev. 14(1), 129–149. Hsieh, Y.H., Ma, S., Hernandez, J.X.V., Lee, V.J. and Lim, W.Y. 2011. Early outbreak of 2009 influenza A (H1N1) in Mexico prior to identification of pH1N1 virus. PLoS One 6(8), e23853. Hu, Z., Tian, X., Lai, R., Ji, C. and Li, X. 2023. Airborne transmission of common swine viruses. Porcine Health Manag. 9(1), 50. Huang, C., Yu, L., Xu, Y., Huang, J., Qin, Y., Guo, X., Zeng, Y., Qin, Y., Ouyang, K., Wei, Z., Huang, W., García-Sastre, A. and Chen, Y. 2024. Long-term co-circulation of multiple influenza A viruses in pigs, Guangxi, China. Emerg. Microbes Infect. 13(1), 2337673. Hussain, M., Galvin, H.D., Haw, T.Y., Nutsford, A.N. and Husain, M. 2017. Drug resistance in influenza A virus: the epidemiology and management. Infect. Drug Resist. 10(1), 121–134. Ison, M.G. 2013. Clinical use of approved influenza antivirals: therapy and prophylaxis. Influenza Other Respir. Viruses 7(Suppl 1), 7–13. Jameson, J., Cruz, J. and Ennis, F.A. 1998. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 72(11), 8682–8689. Jhung, M.A., Swerdlow, D., Olsen, S.J., Jernigan, D., Biggerstaff, M., Kamimoto, L., Kniss, K., Reed, C., Fry, A., Brammer, L., Gindler, J., Gregg, W.J., Bresee, J. and Finelli, L. 2011. Epidemiology of 2009 pandemic influenza A (H1N1) in the United States. Clin. Infect. Dis. 52(Suppl 1), S13–S26. Khatri, M., Dwivedi, V., Krakowka, S., Manickam, C., Ali, A., Wang, L., Qin, Z., Renukaradhya, G.J. and Lee, C.W. 2010. Swine influenza H1N1 virus induces acute inflammatory immune responses in pig lungs: a potential animal model for human H1N1 influenza virus. J. Virol. 84(21), 11210–11218. Kilbourne, E.D. 2006. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 12(1), 9–14. Komadina, N., McVernon, J., Hall, R. and Leder, K. 2014. A historical perspective of influenza A(H1N2) virus. Emerg. Infect. Dis. 20(1), 6–12. Kosik, I. and Yewdell, J.W. 2019. Influenza hemagglutinin and neuraminidase: Yin⁻Yang proteins coevolving to thwart immunity. Viruses 11(4), 346. Lagan, P., Hamil, M., Cull, S., Hanrahan, A., Wregor, R.M. and Lemon, K. 2024. Swine influenza A virus infection dynamics and evolution in intensive pig production systems. Virus Evol. 10(1), veae017. Lessler, J., Cummings, D.A., Fishman, S., Vora, A. and Burke, D.S. 2007. Transmissibility of swine flu at Fort Dix, 1976. J. R. Soc. Interface 4(15), 755–762. Li, Y. and Robertson, I. 2021. The epidemiology of swine influenza. Anim. Dis. 1(1), 21. Lim, B.H. and Mahmood, T.A. 2011. Influenza A H1N1 2009 (Swine Flu) and pregnancy. J. Obstet. Gynaecol. India 61(4), 386–393. Lopez, J.W. and Woods, G.T. 1987. Response of calves to exposure with swine influenza virus. Am. J. Vet. Res. 48(8), 1264–1268. Lorusso, A., Vincent, A.L., Gramer, M.E., Lager, K.M. and Ciacci-Zanella, J.R. 2013. Contemporary epidemiology of North American lineage triple reassortant influenza A viruses in pigs. Curr. Top. Microbiol. Immunol. 370(1), 113–132. Ludwig, S., Haustein, A., Kaleta, E.F. and Scholtissek, C. 1994. Recent influenza A (H1N1) infections of pigs and turkeys in northern Europe. Virology 202(1), 281–286. Lycett, S.J., Duchatel, F. and Digard, P. 2019. A brief history of bird flu. Philos. Trans. R Soc. Lond. B Biol. Sci. 374(1775), 20180257. Ma, W. 2020. Swine influenza virus: current status and challenge. Virus Res. 288(1), 198118. Mastin, A., Alarcon, P., Pfeiffer, D., Wood, J., Williamson, S., Brown, I. and Wieland, B. 2011. Prevalence and risk factors for swine influenza virus infection in the English pig population. PLoS Curr. 3(1), RRN1209. McLean, R.K. and Graham, S.P. 2022. The pig as an amplifying host for new and emerging zoonotic viruses. One Health 14(1), 100384. Mena, I., Nelson, M.I., Quezada-Monroy, F., Dutta, J., Cortes-Fernández, R., Lara-Puente, J.H., Castro-Peralta, F., Cunha, L.F., Trovão, N.S., Lozano-Dubernard, B., Rambaut, A., van Bakel, H. and García-Sastre, A. 2016. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5(1), e16777. Mettelman, R.C., Allen, E.K. and Thomas, P.G. 2022. Mucosal immune responses to infection and vaccination in the respiratory tract. Immunity 55(5), 749–780. Mo, J.S., Abente, E.J., Perez, M.C., Sutton, T.C., Cowan, B., Ferreri, L.M., Geiger, G., Gauger, P.C., Perez, D.R., Baker, A.L.V. and Rajao, D.S. 2022. Transmission of human influenza A virus in pigs selects for adaptive mutations on the HA Gene. J. Virol. 96(22), e0148022. Moghadami, M. 2017. A narrative review of influenza: a seasonal and pandemic disease. Iran. J. Med. Sci. 42(1), 2–13. Monath, T.P. 2013. Vaccines against diseases transmitted from animals to humans: a one health paradigm. Vaccine 31(46), 5321–5338. Moncion, K., Young, K., Tunis, M., Rempel, S., Stirling, R. and Zhao, L. 2019. Effectiveness of hand hygiene practices in preventing influenza virus infection in the community setting: a systematic review. Can. Commun. Dis. Rep. 45(1), 12–23. Nelson, M.I. and Worobey, M. 2018. Origins of the 1918 pandemic: revisiting the swine “mixing vessel” hypothesis. Am. J. Epidemiol. 187(12), 2498–2502. Ng, C., Ye, L., Noorduyn, S.G., Hux, M., Thommes, E., Goeree, R., Ambrose, A., Andrew, M.K., Hatchette, T., Boivin, G., Bowie, W., ElSherif, M., Green, K., Johnstone, J., Katz, K., Leblanc, J., Loeb, M., MacKinnon-Cameron, D., McCarthy, A., McElhaney, J., McGeer, A., Poirier, A., Powis, J., Richardson, D., Sharma, R., Semret, M., Smith, S., Smyth, D., Stiver, G., Trottier, S., Valiquette, L., Webster, D., McNeil, S.A. and Serious Outcomes Surveillance Network of the Canadian Immunization Research Network (CIRN) Investigators; Toronto Invasive Bacterial Diseases Network (TIBDN) Investigators. 2018. Resource utilization and cost of influenza requiring hospitalization in Canadian adults: a study from the serious outcomes surveillance network of the Canadian Immunization Research Network. Influenza Other Respir. Viruses 12(2), 232–240. Njabo, K.Y., Fuller, T.L., Chasar, A., Pollinger, J.P., Cattoli, G., Terregino, C., Monne, I., Reynes, J.M., Njouom, R. and Smith, T.B. 2012. Pandemic A/H1N1/2009 influenza virus in swine, Cameroon, 2010. Vet. Microbiol. 156(1–2), 189–192. Noor, A. and Gradidge, E. 2018. A case of Reye syndrome caused by influenza A virus. Ochsner. J. 18(4), 425–427. Nuwarda, R.F., Alharbi, A.A. and Kayser, V. 2021. An overview of influenza viruses and vaccines. Vaccines (Basel) 9(9), 1032. Ohanu, M.E., Olusina, D.B., Eni, A.O., Aguwa, E.N. and Chukwuka, C.J. 2019. Influenza A viruses: current perspectives on swine flu virus. Int. J. Med. Health Dev. 24(1), 1–8. Ohshima, N., Iba, Y., Kubota-Koketsu, R., Asano, Y., Okuno, Y. and Kurosawa, Y. 2011. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J. Virol. 85(21), 11048–11057. Olsen, C.W., Brammer, L., Easterday, B.C., Arden, N., Belay, E., Baker, I. and Cox, N.J. 2002. Serologic evidence of H1 swine Influenza virus infection in swine farm residents and employees. Emerg. Infect. Dis. 8(8), 814–819. O’Riordan, S., Barton, M., Yau, Y., Read, S.E., Allen, U. and Tran, D. 2010. Risk factors and outcomes among children admitted to hospital with pandemic H1N1 influenza. CMAJ 182(1), 39–44. Ouchi, A., Nerome, K., Kanegae, Y., Ishida, M., Nerome, R., Hayashi, K., Hashimoto, T., Kaji, M., Kaji, Y. and Inaba, Y. 1996. Large outbreak of swine influenza in southern Japan caused by reassortant (H1N2) influenza viruses: its epizootic background and characterization of the causative viruses. J. Gen. Virol. 77(Pt 8), 1751–1759. Pantaleo, G., Correia, B., Fenwick, C., Joo, V.S. and Perez, L. 2022. Antibodies to combat viral infections: development strategies and progress. Nat. Rev. Drug Discov. 21(9), 676–696. Peiris, J.S., de Jong, M.D. and Guan, Y. 2007. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 20(2), 243–267. Peiris, J.S., Guan, Y., Markwell, D., Ghose, P., Webster, R.G. and Shortridge, K.F. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75(20), 9679–9686. Peteranderl, C., Herold, S. and Schmoldt, C. 2016. Human influenza virus infections. Semin. Respir. Crit. Care Med. 37(4), 487–500. Petro-Turnquist, E., Pekarek, M.J. and Weaver, E.A. 2024. Swine influenza A virus: challenges and novel vaccine strategies. Front. Cell. Infect. Microbiol. 14(1), 1336013. Poulson, R.L., Tompkins, S.M., Berghaus, R.D., Brown, J.D. and Stallknecht, D.E. 2016. Environmental stability of swine and human pandemic influenza viruses in water under variable conditions of temperature, salinity, and pH. Appl. Environ. Microbiol. 82(13), 3721–3726. Rak, A., Isakova-Sivak, I. and Rudenko, L. 2023. Nucleoprotein as a promising antigen for broadly protective influenza vaccines. Vaccines 11(12), 1747. Ravina, R., Dalal, A., Mohan, H., Prasad, M. and Pundir, C.S. 2020. Detection methods for influenza A H1N1 virus with special reference to biosensors: a review. Biosci. Rep. 40(2), BSR20193852. Rewar, S., Mirdha, D. and Rewar, P. 2015. Treatment and prevention of pandemic H1N1 influenza. Ann. Glob. Health 81(5), 645–653. Richard, M. and Fouchier, R.A. 2016. Influenza A virus transmission via respiratory aerosols or droplets as it relates to pandemic potential. FEMS Microbiol. Rev. 40(1), 68–85. Richt, J.A., Lekcharoensuk, P., Lager, K.M., Vincent, A.L., Loiacono, C.M., Janke, B.H., Wu, W.H., Yoon, K.J., Webby, R.J., Solórzano, A. and García-Sastre, A. 2006. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J. Virol. 80(22), 11009–11018. Robinson, J.L., Lee, B.E., Patel, J., Bastien, N., Grimsrud, K., Seal, R.F., King, R., Marshall, F. and Li, Y. 2007. Swine influenza (H3N2) infection in a child and possible community transmission, Canada. Emerg. Infect. Dis. 13(12), 1865–1870. Rovida, F., Piralla, A., Marzani, F.C., Moreno, A., Campanini, G., Mojoli, F., Pozzi, M., Girello, A., Chiapponi, C., Vezzoli, F., Prati, P., Percivalle, E., Pavan, A., Gramegna, M., Iotti, G.A. and Baldanti, F. 2016. Swine influenza A (H1N1) virus (SIV) infection requiring extracorporeal life support in an immunocompetent adult patient with indirect exposure to pigs, Italy, October 2016. Euro Surveill. 22(5), 30456. Ryt-Hansen, P., Pedersen, A.G., Larsen, I., Krog, J.S., Kristensen, C.S. and Larsen, L.E. 2019. Acute Influenza A virus outbreak in an enzootic infected sow herd: Impact on viral dynamics, genetic and antigenic variability and effect of maternally derived antibodies and vaccination. PLoS One 14(11), e0224854. Said, A., Lange, E., Beer, M., Damiani, A. and Osterrieder, N. 2013. Recombinant equine herpesvirus 1 (EHV-1) vaccine protects pigs against challenge with influenza A(H1N1)pmd09. Virus Res. 173(2), 371–376. Salvesen, H.A. and Whitelaw, C.B.A. 2021. Current and prospective control strategies of influenza A virus in swine. Porcine Health Manag. 7(1), 23. Sarli, G., D’Annunzio, G., Gobbo, F., Benazzi, C. and Ostanello, F. 2021. The role of pathology in the diagnosis of swine respiratory disease. Vet. Sci. 8(11), 256. Schmidt, C.W. 2009. Swine CAFOs and novel H1N1 flu: separating facts from fears. Environ. Health Perspect. 117(9), A394–A401. Schnitzler, S.U. and Schnitzler, P. 2009. An update on swine-origin influenza virus A/H1N1: a review. Virus Genes 39(3), 279–292. Schorr, E., Wentworth, D. and Hinshaw, V.S. 1994. Use of polymerase chain reaction to detect swine influenza virus in nasal swab specimens. Am. J. Vet. Res. 55(7), 952–956. Short, K.R., Richard, M., Verhagen, J.H., van Riel, D., Schrauwen, E.J., van den Brand, J.M., Mänz, B., Bodewes, R. and Herfst, S. 2015. One health, multiple challenges: the inter-species transmission of influenza A virus. One Health 1(1), 1–13. Shurson, G.C., Urriola, P.E. and Schroeder, D.C. 2023. Biosecurity and mitigation strategies to control swine viruses in feed ingredients and complete feeds. Animals 13(14), 2375. Smith, R.D., Keogh-Brown, M.R., Barnett, T. and Tait, J. 2009. The economy-wide impact of pandemic influenza on the UK: a computable general equilibrium modelling experiment. BMJ 339(1), b4571. Smyk, J.M., Szydłowska, N., Szulc, W. and Majewska, A. 2022. Evolution of influenza viruses-drug resistance, treatment options, and prospects. Int. J. Mol. Sci. 23(20), 12244. Sreenivasan, C.C., Thomas, M., Kaushik, R.S., Wang, D. and Li, F. 2019. Influenza A in bovine species: a narrative literature review. Viruses 11(6), 561. Sreta, D., Kedkovid, R., Tuamsang, S., Kitikoon, P. and Thanawongnuwech, R. 2009. Pathogenesis of swine influenza virus (Thai isolates) in weanling pigs: an experimental trial. Virol. J. 6(1), 34. Staneková, Z. and Varečková, E. 2010. Conserved epitopes of influenza A virus inducing protective immunity and their prospects for universal vaccine development. Virol. J. 7(1), 351. Suarez, D.L., Perdue, M.L., Cox, N., Rowe, T., Bender, C., Huang, J. and Swayne, D.E. 1998. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J. Virol. 72(8), 6678–6688. Takemae, N., Nguyen, T., Ngo, L.T., Hiromoto, Y., Uchida, Y., Pham, V.P., Kageyama, T., Kasuo, S., Shimada, S., Yamashita, Y., Goto, K., Kubo, H., Le, V.T., Van Vo, H., Do, H.T., Nguyen, D.H., Hayashi, T., Matsuu, A. and Saito, T. 2013. Antigenic variation of H1N1, H1N2 and H3N2 swine influenza viruses in Japan and Vietnam. Arch. Virol. 158(4), 859–876. Taubenberger, J.K. and Kash, J.C. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7(6), 440–451. van de Sandt, C.E., Kreijtz, J.H. and Rimmelzwaan, G.F. 2012. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 4(9), 1438–1476. van Diemen, P.M., Byrne, A.M.P., Ramsay, A.M., Watson, S., Nunez, A., V Moreno, A., Chiapponi, C., Foni, E., Brown, I.H., Brookes, S.M. and Everett, H.E. 2023. Interspecies transmission of swine influenza A viruses and human seasonal vaccine-mediated protection investigated in ferret model. Emerg. Infect. Dis. 29(9), 1798–1807. Vasoo, S., Stevens, J. and Singh, K. 2009. Rapid antigen tests for diagnosis of pandemic (Swine) influenza A/H1N1. Clin. Infect. Dis. 49(7), 1090–1093. Vincent, A.L., Ma, W., Lager, K.M., Janke, B.H. and Richt, J.A. 2008. Swine influenza viruses a North American perspective. Adv. Virus Res. 72(1), 127–154. Wang, C., Yu, H., Horby, P.W., Cao, B., Wu, P., Yang, S., Gao, H., Li, H., Tsang, T.K., Liao, Q., Gao, Z., Ip, D.K., Jia, H., Jiang, H., Liu, B., Ni, M.Y., Dai, X., Liu, F., Van Kinh, N., Liem, N.T., Hien, T.T., Li, Y., Yang, J., Wu, J.T., Zheng, Y., Leung, G.M., Farrar, J.J., Cowling, B.J., Uyeki, T.M. and Li, L. 2014. Comparison of patients hospitalized with influenza A subtypes H7N9, H5N1, and 2009 pandemic H1N1. Clin. Infect. Dis. 58(8), 1095–1103. Webby, R.J., Swenson, S.L., Krauss, S.L., Gerrish, P.J., Goyal, S.M. and Webster, R.G. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 74(18), 8243–8251. Webster, R.G. and Govorkova, E.A. 2014. Continuing challenges in influenza. Ann. N. Y. Acad. Sci. 1323(1), 115–139. Weingartl, H.M., Albrecht, R.A., Lager, K.M., Babiuk, S., Marszal, P., Neufeld, J., Embury-Hyatt, C., Lekcharoensuk, P., Tumpey, T.M., García-Sastre, A. and Richt, J.A. 2009. Experimental infection of pigs with the human 1918 pandemic influenza virus. J. Virol. 83(9), 4287–4296. White, M.R., Doss, M., Boland, P., Tecle, T. and Hartshorn, K.L. 2008. Innate immunity to influenza virus: implications for future therapy. Expert Rev. Clin. Immunol. 4(4), 497–514. Yu, H., Hua, R.H., Zhang, Q., Liu, T.Q., Liu, H.L., Li, G.X. and Tong, G.Z. 2008. Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J. Clin. Microbiol. 46(3), 1067–1075. Zeller, M.A., Ma, J., Wong, F.Y., Tum, S., Hidano, A., Holt, H., Chhay, T., Sorn, S., Koeut, D., Seng, B., Chao, S., Ng, G.G.K., Yan, Z., Chou, M., Rudge, J.W., Smith, G.J.D. and Su, Y.C.F. 2023. The genomic landscape of swine influenza A viruses in Southeast Asia. Proc. Natl. Acad. Sci. U S A 120(33), e2301926120. Zhang, H., Li, X., Ma, R., Li, X., Zhou, Y., Dong, H., Li, X., Li, Q., Zhang, M., Liu, Z., Wei, B., Cui, M., Wang, H., Gao, J., Yang, H., Hou, P., Miao, Z. and Chai, T. 2013. Airborne spread and infection of a novel swine-origin influenza A (H1N1) virus. Virol. J. 10(1), 204. Zhang, J. and Gauger, P.C. 2014. Isolation of swine influenza virus in cell cultures and embryonated chicken eggs. Methods Mol. Biol. 1161(1), 265–276. Zhang, Y., Sun, W., Svendsen, E.R., Tang, S., MacIntyre, R.C., Yang, P., Zhang, D. and Wang, Q. 2015. Do corticosteroids reduce the mortality of influenza A (H1N1) infection? A meta-analysis. Crit. Care 19(1), 46. Zhang, Y., Xu, C., Zhang, H., Liu, G.D., Xue, C. and Cao, Y. 2019. Targeting hemagglutinin: approaches for broad protection against the influenza A virus. Viruses 11(5), 405. | ||
| How to Cite this Article |
| Pubmed Style Meles DK, Khairullah AR, Rimayanti R, Mustofa I, Wurlina W, Utama S, Lestari TD, Mulyati S, Ahmad RZ, Moses IB, Wibowo S, Kusala MKJ, Wardhani BWK, Fauziah I, Kurniasih DAA, Anggraini L, Ekawasti F, Akintunde AO. The global burden of swine influenza and its mitigation. Open Vet. J.. 2025; 15(5): 1866-1879. doi:10.5455/OVJ.2025.v15.i5.3 Web Style Meles DK, Khairullah AR, Rimayanti R, Mustofa I, Wurlina W, Utama S, Lestari TD, Mulyati S, Ahmad RZ, Moses IB, Wibowo S, Kusala MKJ, Wardhani BWK, Fauziah I, Kurniasih DAA, Anggraini L, Ekawasti F, Akintunde AO. The global burden of swine influenza and its mitigation. https://www.openveterinaryjournal.com/?mno=237319 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i5.3 AMA (American Medical Association) Style Meles DK, Khairullah AR, Rimayanti R, Mustofa I, Wurlina W, Utama S, Lestari TD, Mulyati S, Ahmad RZ, Moses IB, Wibowo S, Kusala MKJ, Wardhani BWK, Fauziah I, Kurniasih DAA, Anggraini L, Ekawasti F, Akintunde AO. The global burden of swine influenza and its mitigation. Open Vet. J.. 2025; 15(5): 1866-1879. doi:10.5455/OVJ.2025.v15.i5.3 Vancouver/ICMJE Style Meles DK, Khairullah AR, Rimayanti R, Mustofa I, Wurlina W, Utama S, Lestari TD, Mulyati S, Ahmad RZ, Moses IB, Wibowo S, Kusala MKJ, Wardhani BWK, Fauziah I, Kurniasih DAA, Anggraini L, Ekawasti F, Akintunde AO. The global burden of swine influenza and its mitigation. Open Vet. J.. (2025), [cited January 25, 2026]; 15(5): 1866-1879. doi:10.5455/OVJ.2025.v15.i5.3 Harvard Style Meles, D. K., Khairullah, . A. R., Rimayanti, . R., Mustofa, . I., Wurlina, . W., Utama, . S., Lestari, . T. D., Mulyati, . S., Ahmad, . R. Z., Moses, . I. B., Wibowo, . S., Kusala, . M. K. J., Wardhani, . B. W. K., Fauziah, . I., Kurniasih, . D. A. A., Anggraini, . L., Ekawasti, . F. & Akintunde, . A. O. (2025) The global burden of swine influenza and its mitigation. Open Vet. J., 15 (5), 1866-1879. doi:10.5455/OVJ.2025.v15.i5.3 Turabian Style Meles, Dewa Ketut, Aswin Rafif Khairullah, Rimayanti Rimayanti, Imam Mustofa, Wurlina Wurlina, Suzanita Utama, Tita Damayanti Lestari, Sri Mulyati, Riza Zainuddin Ahmad, Ikechukwu Benjamin Moses, Syahputra Wibowo, Muhammad Khaliim Jati Kusala, Bantari Wisynu Kusuma Wardhani, Ima Fauziah, Dea Anita Ariani Kurniasih, Lili Anggraini, Fitrine Ekawasti, and Adeyinka Oye Akintunde. 2025. The global burden of swine influenza and its mitigation. Open Veterinary Journal, 15 (5), 1866-1879. doi:10.5455/OVJ.2025.v15.i5.3 Chicago Style Meles, Dewa Ketut, Aswin Rafif Khairullah, Rimayanti Rimayanti, Imam Mustofa, Wurlina Wurlina, Suzanita Utama, Tita Damayanti Lestari, Sri Mulyati, Riza Zainuddin Ahmad, Ikechukwu Benjamin Moses, Syahputra Wibowo, Muhammad Khaliim Jati Kusala, Bantari Wisynu Kusuma Wardhani, Ima Fauziah, Dea Anita Ariani Kurniasih, Lili Anggraini, Fitrine Ekawasti, and Adeyinka Oye Akintunde. "The global burden of swine influenza and its mitigation." Open Veterinary Journal 15 (2025), 1866-1879. doi:10.5455/OVJ.2025.v15.i5.3 MLA (The Modern Language Association) Style Meles, Dewa Ketut, Aswin Rafif Khairullah, Rimayanti Rimayanti, Imam Mustofa, Wurlina Wurlina, Suzanita Utama, Tita Damayanti Lestari, Sri Mulyati, Riza Zainuddin Ahmad, Ikechukwu Benjamin Moses, Syahputra Wibowo, Muhammad Khaliim Jati Kusala, Bantari Wisynu Kusuma Wardhani, Ima Fauziah, Dea Anita Ariani Kurniasih, Lili Anggraini, Fitrine Ekawasti, and Adeyinka Oye Akintunde. "The global burden of swine influenza and its mitigation." Open Veterinary Journal 15.5 (2025), 1866-1879. Print. doi:10.5455/OVJ.2025.v15.i5.3 APA (American Psychological Association) Style Meles, D. K., Khairullah, . A. R., Rimayanti, . R., Mustofa, . I., Wurlina, . W., Utama, . S., Lestari, . T. D., Mulyati, . S., Ahmad, . R. Z., Moses, . I. B., Wibowo, . S., Kusala, . M. K. J., Wardhani, . B. W. K., Fauziah, . I., Kurniasih, . D. A. A., Anggraini, . L., Ekawasti, . F. & Akintunde, . A. O. (2025) The global burden of swine influenza and its mitigation. Open Veterinary Journal, 15 (5), 1866-1879. doi:10.5455/OVJ.2025.v15.i5.3 |