| Research Article | ||
Open Vet. J.. 2025; 15(7): 2948-2958 Open Veterinary Journal, (2025), Vol. 15(7): 2948-2958 Research Article Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in IraqDoaa Sabri Resen and Noor Idan Jarad*Department of Veterinary Microbiology, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq *Corresponding Author: Noor Idan Jarad, Department of Veterinary mMicrobiology, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq. Email: noor.jarad [at] qu.edu.iq Submitted: 16/02/2025 Revised: 16/06/2025 Accepted: 21/06/2025 Published: 31/07/2025 © 2025 Open Veterinary Journal
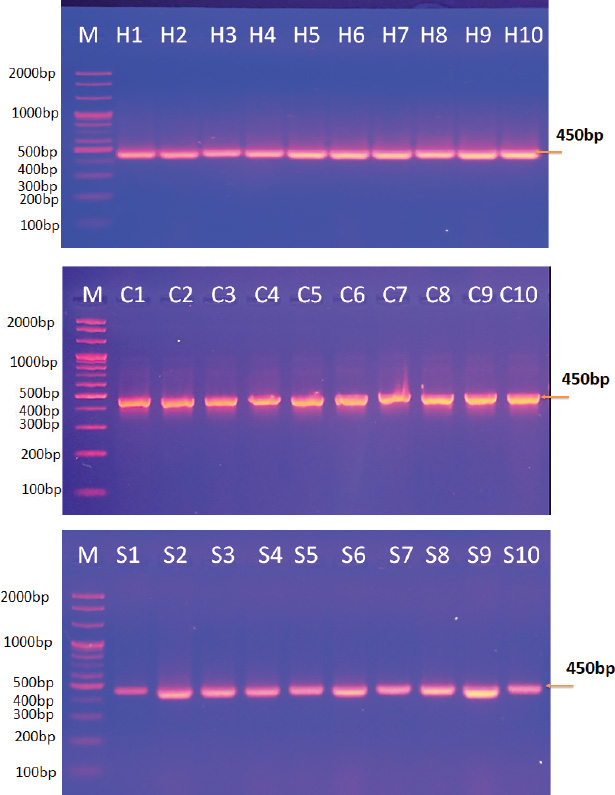
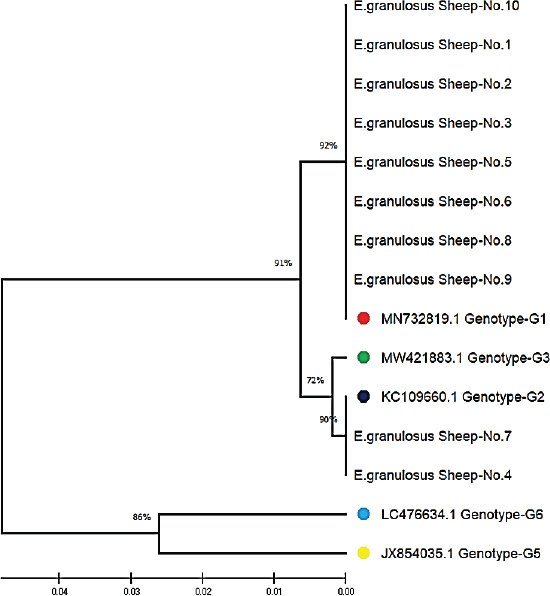
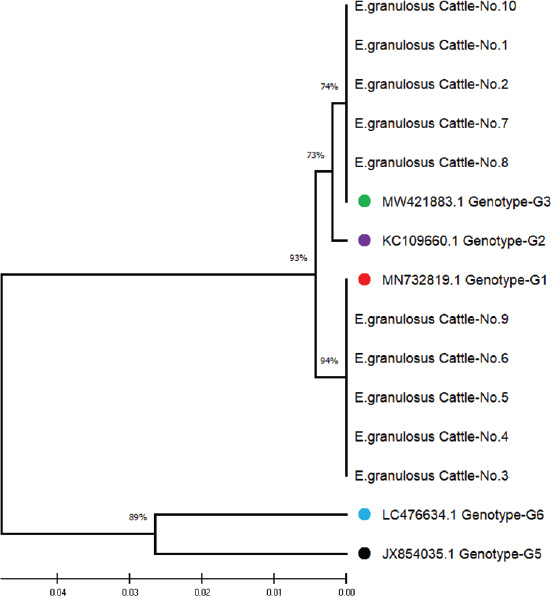
ABSTRACTBackground: Cystic echinococcosis (CE) is a parasitic infection caused by the larval form of Echinococcus granulosus (a member of the Taeniidae family). The infection is spread by fecal-oral contact with infected domestic dogs. CE threatens human health and destroys animal production all over the world. Echinococcus granulosus genotypes need to be correctly identified for better control and prevention. Aim: This study isolated the genotypes of E. granulosus from human, sheep, and cattle samples collected in Iraq using molecular and phylogenetic methods. Methods: Thirty hydatid cyst samples were collected: 10 from human patients in the hospital and 20 from sheep and cattle slaughtered in Al-Diwaniyah province. DNA was extracted using a commercial kit, and PCR amplification of the mitochondrial Cox1 gene was conducted. The amplified products were sequenced, and the sequences were studied using phylogeny. Results: The findings showed three genotypes of E. granulosus (G1, G2, and G3). G1 was more common in human isolates (98.40%–99.70% homology), G1 and G2 were also found in sheep (98.34%–99.70% homology), and G1 and G3 in cattle (98.40%–99.75% homology). Phylogenetic analysis identified a clear clustering of genotypes that belonged to the same host. The G1 genotype was detected as the most common strain that infects people, sheep, and cattle in Iraq. It is highly zoonotic and has the highest economic losses. Conclusion: This study confirmed the presence of three genotypes of E. granulosus in Iraq: G1, G2, and G3. G1 was most common in all hosts, particularly in humans. Phylogenetic analysis revealed that each host had closely related strains. G1 poses a serious public health and economic threat due to its high zoonotic potential. Keywords: Cattle, Echinococcus granulosus, Genotype, Human, Iraq, Sheep. IntroductionEchinococcus is a genus of cyclophyllid cestodes. Adult worms live in the small intestines of definitive carnivorous hosts, including dogs, wolves, and hyenas, whereas juveniles, known as metacestodes, reside in the internal organs of various mammalian intermediate hosts. Overall, there are 37 known Echinococcus species, which are mainly defined based on minute morphological differences from the larval stage; however, only 14 species are considered valid due to the availability of molecular data (Vishniakov et al., 2024). Humans also act as intermediate hosts (Widdicombe et al., 2022). Adult Echinococcus worms cause carnivorous Echinococcosis, while larvae cause hydatid disease in humans, which is a worldwide public health hazard. Echinococcosis is classified under the category of “neglected zoonotic diseases” and has an important socioeconomic and public health impact in many parts of the world, particularly in regions with lower socioeconomic status (Vishniakov et al., 2024). Echinococcosis has been a public health issue since the time of Hippocrates; however, the disease has taken an extra dimension, particularly in China, in terms of its global public health importance. Echinococcosis increases the total number of DALYs caused by parasitic diseases by 19.7% in certain countries (Gholami et al., 2021). The two principal and most widely distributed species of Echinococcus are Echinococcus granulosus and Echinococcus multilocularis. One of the unique and interesting biological features of these groups of Echinococcosis is their capacity to grow and survive in multiple mammalian hosts, particularly in extreme climates and environments, in different parts of the world (Oliveira et al., 2022). Concerning the economic impact, losses have been caused concurrently or consequently by infections with Echinococcosis (Xie et al., 2023). This section deals with the global and regional prevalence. Although the disease is endemic throughout the world with considerable variations in terms of intensity and distribution across countries, Echinococcosis is reported in four continents: Latin America, Asia, Eastern Europe, and Africa (Ali et al., 2024). The distribution pattern of human Echinococcosis closely follows the livestock farming distribution, underpinning the extent of direct and indirect contributions to well-being, poverty alleviation, and socioeconomics, predicated by the direct linkage between climate change and ensuing changes in transhumance patterns and frequency (Hogea et al., 2024). Strengthening intestinal health in people may boost the immune system against hydatid cysts (Gholami et al., 2021; Ghazi et al., 2024). It is of practical significance to control Echinococcosis by blocking related factors through interfering with different links in the development of life cycle infection in intermediate hosts, treatment or immunization of definitive hosts, and preventative measures in parasitic survival and development, which is a comprehensive measure requiring long-term efforts (Omondi et al., 2020; Gholami et al., 2021; Janabi, 2021; Derakhshani et al., 2023; Guo et al., 2023; Almashhadany et al., 2024; Kheninef et al., 2024). This study isolated the genotypes of E. granulosus from human, sheep, and cattle samples collected in Iraq using molecular and phylogenetic methods. Materials and MethodsSamplesThirty hydatid cysts were obtained from three hosts, 10 from human patients receiving surgical treatment for cystic Echinococcosis, 10 from sheep, and 10 from cattle at a slaughterhouse in Al-Diwaniyah province, Iraq. The protocolizes was snipped from the cyst fluid as described by Agha (2015) and stored for molecular analysis. Molecular identificationDNA extractionAll samples were extracted from DNA using a Geneaid Genomic DNA Extraction Kit (Taiwan) according to the manufacturer’s instructions. The purity and concentration of the extracted DNA were measured using a Nanodrop spectrophotometer. All the taken DNA was kept at −20°C until analysis. Polymerase chain reactionPrimer from Nikmanesh et al. (2014) PCR amplification of the mitochondrial Cox1 gene. The primer sequence forward was 5’-TTTTTTGGGCATCCTGAGGTTTAT-3’ and the reverse primer sequence 5’-TAAAGAAAGAACATAATGAAAATG-3’. The PCR reactions were run using a Maxime PCR PreMix Kit (iNtRON, Korea) containing Taq DNA polymerase, dNTPs, Tris-HCl buffer (pH 9.0), KCl, MgCl2, stabilizer, and tracking dye. In each 20 µl reaction, 5 µl template DNA, 1 µl of each primer (10 pmol), and 13 µl PCR-compatible water were added. The PCR conditions were as follows: denaturation at 94°C for 5 minutes in a thermocycler (Thermo Fisher Scientific, USA), followed by 35 denaturation cycles of 94°C for 45 seconds, annealing at 50°C for 45 seconds, and extension at 72°C for 45 seconds. The final extension step was performed at 72 °C for 7 minutes, followed by sample storage at 4 °C for analysis. SequencingThe PCR products were analyzed by agarose gel electrophoresis and plotted using UV light. Positive amplicons were sent to Macrogen Company (Korea) for sequencing on an AB DNA sequencing machine. The phylogenetic tree was constructed using MEGA 6.0 software with the use of multiple sequence alignment by ClustalW. Evolutionary relations were calculated using maximum composite likelihood, and a tree of life was established using UPGMA. Ethical approvalThe current research was conducted after receiving ethical approval (1890/28/8/2023) from the Committee for Research Ethics at the College of Veterinary Medicine, University of Al-Qadisiyah, Diwaniyah, Iraq. ResultsPolymerase chain reactionThey examined 30 hydatid cysts from people, sheep, and cattle in Iraq. The researchers could extract DNA from protoscolices from all samples and PCR-amplification of the mitochondrial Cox1 gene. This PCR product was always 450-base-pairs molecular weight and showed E. granulosus DNA in every sample (Fig. 1). Sequencing and multiple alignmentDNA was sequenced from local E. granulosus isolates from humans, sheep, and cattle using the mitochondrial Cox1 gene. We subjected sequences to multiple alignments against E. granulosus genotypes in the NCBI-BLAST database using ClustalW. Phylogenetic reconstruction using the maximum composite likelihood algorithm to build a UPGMA tree in MEGA 6.0. The 10 human isolates (No. 1–10) exhibited strong genetic similarity to the G1 genotype according to NCBI-BLAST results. The genetic variation ranged from 0.05 to 0.01, and the sequence identity was between 98.40% and 99.70%. In sheep, eight isolates (No. 1, 2, 3, 5, 6, 8, 9, and 10) matched the G1 genotype with 98.34%–99.65% similarity, whereas two (No. 4 and 7) matched G2 with 99.30%–99.70%. Among the cattle samples, five (No. 3, 4, 5, 6, and 9) were identified as G1 (98.40%–99.75%), and the remaining five (No. 1, 2, 7, 8, and 10) matched G3 (98.65%–99.50%).
Fig. 1. Agarose gel electrophoresis image showing the PCR product analysis of Echinococcus granulosus mitochondrial cytochrome oxidase subunit 1 (COX1) gene from extracted DNA of hydatid cyst fluid of humans (H1-H10: positive samples), sheep (S1-S10: Positive samples), and cattle (C1-C10: Positive samples). M: marker (2,000-100bp).
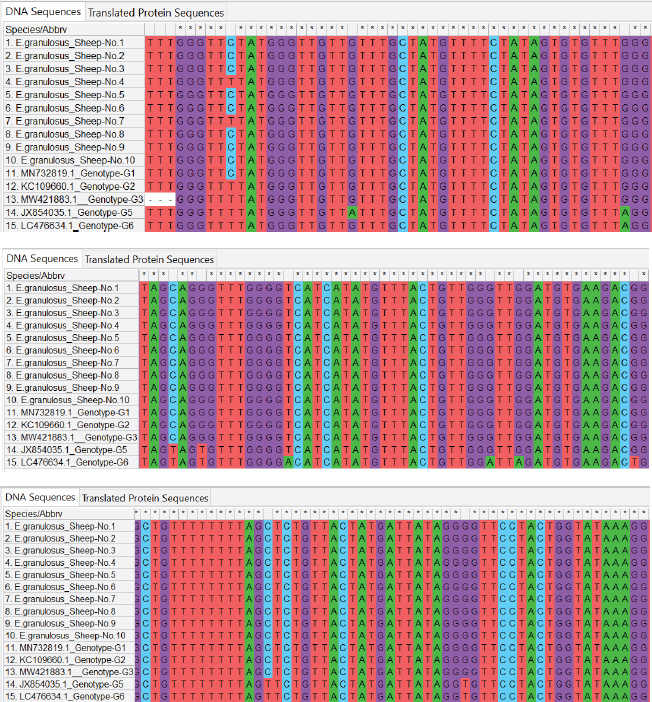
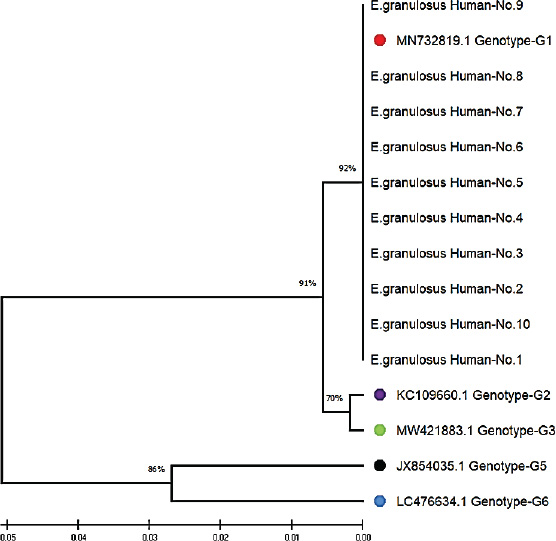
Fig. 2. ClustalW multiple sequence alignment of mitochondrial Cox1 gene partial sequences from local Echinococcus granulosus human isolates. This comparison aligned the sequences with the associated E. granulosus genotypes from the NCBI-BLAST database. The alignment indicates areas of homology (marked with stars (*), corresponding to conserved sequences, and genetic divergence between the nucleotide sequences of the human isolates. These results illustrate the genetic diversity and conserved elements of the mitochondrial Cox1 gene of E. granulosus. All local isolates of E. granulosus were added to NCBI GenBank under the accession numbers PP979853.1–PP979857.1. These data show that human, sheep, and cattle hosts in Iraq were dominant in G1, G2, and G3 (Figs. 2–4). Phylogenetic analysisPhylogenetic reconstruction showed that human, sheep, and cattle isolates were tightly clustered around their genotypes, with different branches on the tree. Isolates from humans were closely related to the G1 strain, whereas those from sheep were classified as G1 and G2. Isolates from cattle showed the same split, with some grouped together with G1 and others grouped with G3. These results were supported by multiple sequence alignments, showing that the local isolates shared similarities and differences in genetic material with reference sequences.
Fig. 3. ClustalW multiple sequence alignment of partial mitochondrial Cox1 gene sequences from local Echinococcus granulosus sheep isolates. This comparison aligned the sequences with the associated E. granulosus genotypes from the NCBI-BLAST database. The alignment indicates areas of homology (marked with stars (*), corresponding to conserved sequences, and genetic divergence between the nucleotide sequences of the sheep isolates. These results illustrate the genetic diversity and conserved elements of the mitochondrial Cox1 gene of E. granulosus.
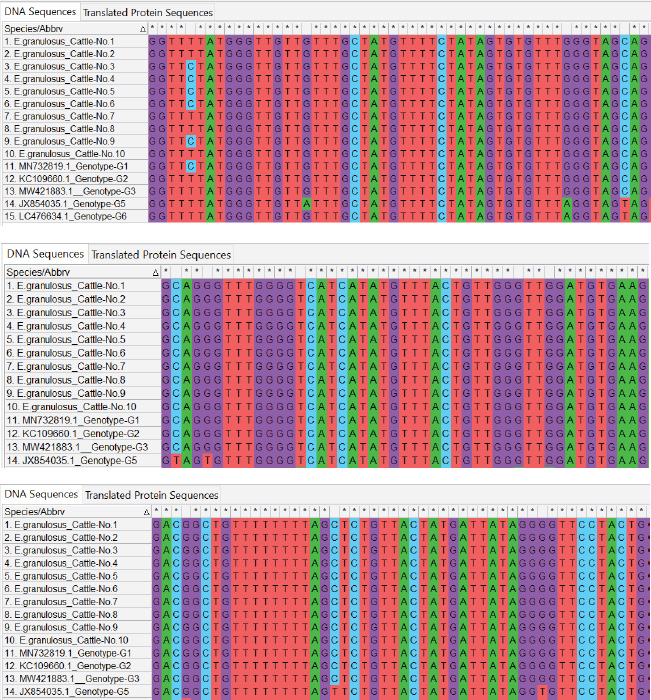
Fig. 4. ClustalW multiple sequence alignment of partial mitochondrial Cox1 gene sequences from local Echinococcus granulosus cattle isolates. This comparison aligned the sequences with the associated E. granulosus genotypes from the NCBI-BLAST database. The alignment indicates areas of homology (marked with stars (*), corresponding to conserved sequences, and genetic divergence between the nucleotide sequences of the cattle isolates. These results illustrate the genetic diversity and conserved elements of the mitochondrial Cox1 gene of E. granulosus. This research also managed to deposit sequences of the local isolates in the NCBI GenBank database, with accession numbers PQ661280, PQ661281, PQ661282, PQ661283, and PQ661284 for sheep, human, and cattle isolates (Figs. 5–7). DiscussionThe current study genotyped E. granulosus strains from humans, sheep, and cattle in Iraq and shed light on the molecular epidemiology of this parasite. The G1 genotype prevails in human isolates (98.40%–99.70% homology), and this has been found in a few international studies. Shahabi et al. (2021) declared G1 to be the predominant genotype in humans in Turkey and Iran and reported that it is of zoonotic importance and widespread incidence. Similarly, Hodžić et al. (2022) identified G1 as the most prevalent genotype in Bosnia and Herzegovina, supporting its worldwide dominance. This dominant position results from close contact between humans and infected animals, notably sheep, which serve as the main intermediate hosts for G1.
Fig. 5. Phylogenetic tree analysis based on the partial mitochondrial Cox1 sequence of Echinococcus granulosus from local human samples (color-filled circles). This is based on the evolutionary distances and Maximum Composite Likelihood method (UPGMA tree) in (MEGA 6.0 version). By contrast, research by Celik et al. (2024) derived both the G1 and G3 genotypes from human isolates in Turkey, with regional differences in genotype distribution. Moreover, in Bolivia Ali et al. (2020) recorded an expanded range of genotypes from G3 to G7, indicating geographical variation across E. granulosus transmission cycles. These results suggest that although G1 is the global champion, other genotypes, such as G3, also contribute to human infection in certain areas. The identification of the G1 (98.34%–99.65% homology) and G2 (99.30%–99.70%) genotypes in sheep isolates concurs with observations from Turkey (Talladaçalşr et al., 2022) reported G1 as the most common genotype and G2 as a small strain. Similarly, Aziz et al. discovered a preference for the G1 and G3 genotypes in sheep from the US, and these genotypes are common in sheep populations around the world (Jesudoss Chelladurai et al., 2024).
Fig. 6. Phylogenetic tree analysis based on the partial mitochondrial Cox1 sequence of Echinococcus granulosus from local sheep samples (color-filled circles). This is based on the evolutionary distances and Maximum Composite Likelihood method (UPGMA tree) in (MEGA 6.0 version). Interestingly, Pereira et al. (2022) discovered the difference in the expression of genes for immune modulation between cattle and sheep cysts that may influence genotype diversity between hosts. In addition, Aziz et al. (2022) conducted studies in Kurdistan, Iraq, and also detected a greater frequency of hydatid cysts in sheep (66.7%), providing evidence for host-specific dynamics in infection behavior. These results indicate that sheep are primary intermediate hosts during the life course of G1 and related genotypes. In the cattle, the G1 (98.40%–99.75%) and G3 (98.65%–99.50%) genotypes were identified, in accordance with various studies. Celik et al. (2024) confirmed the genotypes of G1 and G3 in cow isolates from Turkey as both hosts and intermediaries. In the United States, Jesudoss Chelladurai et al. (2024) verified that the G1 and G3 genotypes in cattle are endemic to domestic dog ruminant cycles (Jesudoss Chelladurai et al., 2024). Contrastingly, Gholami et al. (2021) reported the discovery of G5 (buffalo strain) in sheep and goats in Iran, implying that host- and environment-driven genotype variations occur regionally. Additionally, Kheninef et al. (2024) observed intragenotype variation in the G1 and G3 genotypes in Algeria, indicating genetic variation even within the dominant strains. These variations could be accounted for by regional variations in genotype incidence and pathogenicity (Karim et al., 2019).
Fig. 7. Phylogenetic tree analysis based on the mitochondrial Cox1 gene partial sequence used for E. granulosus from local cattle samples (color-filled circles). This is based on the evolutionary distances and Maximum Composite Likelihood method (UPGMA tree) in (MEGA 6.0 version). Because G1 is dominant in isolates from humans, sheep, and cattle, its zoonotic importance and cross-host adaptability are clear. Studies by Omondi et al. (2020) from Kenya and Guo et al. (2023) from China also reported G1 as the dominant genotype among several host species, confirming its international validity. However, G2 and G3, in particular hosts, indicate that region-wide epidemiological research is needed to get a better handle on how genotypes are distributed and how they shift in transmission (Janabi, 2021). Genetic variations among E. granulosus isolates are relevant for disease surveillance. Pereira et al. (2022) demonstrated how genotype-dependent immune regulation in host-specific cells could be manipulated for vaccine and therapy development. What’s more, the introduction of genotypes such as G5 by Gholami et al. (2021) also has people worried about E. granulosus’s adaptation to novel hosts and environments that must constantly be monitored. ConclusionThis study confirmed the presence of three genotypes of E. granulosus in Iraq: G1, G2, and G3. G1 was most common in all hosts, particularly in humans. Phylogenetic analysis revealed that each host had closely related strains. G1 poses a serious public health and economic threat due to its high zoonotic potential. AcknowledgmentNone. FundingThis work was self-funded by the authors. Authors’ contributionsAll authors have participated equally in the research. Conflict of interestThe authors declare no conflict of interest. Data availabilityAll data can be provided upon request. ReferencesAgha, S.A.F.A. 2015. Genotyping of cystic Echinococcosis isolates from human and animals clinical samples. MSc Thesis, College of Veterinary Medicine, University of Al-Qadisiyah. Ali, R., Nazeer, S., Elahi, M.M.S., Idu, E.G., Zhang, H., Mahmoudvand, H., Khan, S.N. and Yang, J. 2024. Global distribution and definitive host range of Echinococcus species and genotypes: a systematic review. Vet. Parasitol. 331, 110273; doi: 10.1016/j.vetpar.2024.110273. Almashhadany, D., Aziz Jamil Alani, A., Ahmed Dhiab, A., AbdulMona M. Zainel, M. and Talal Abdulrahman, T. 2024. Public health significance of human toxoplasmosis. IntechOpen; doi: 10.5772/intechopen.114338. Aziz, H.M., Hama, A.A. and Hama Salih, M.A. 2022. An epidemiological study of hydatid cyst of Echinococcus granulosus in Sulaimani, Iraq. Ann. Parasitol. 68(2), 241–246. Celik, F., Selcuk, M. A., Kilinc, S. G., Kesik, H. K., Ahmed, H., Wang, Y., Simsek, S. and Cao, J. 2024. Molecular discrimination of G1 and G3 genotypes of Echinococcus granulosus sensu stricto obtained from human, cattle, and sheep using the mitochondrial NADH dehydrogenase subunit 5 marker. Acta Trop. 252, 107124. Derakhshani, A., Mousavi, S.M., Rezaei, M., Afgar, A., Keyhani, A.R. and Mohammadi, M.A., Dabiri, S. and Fasihi Harandi, M. 2023. Natural history of Echinococcus granulosus microcyst development in long-term in vitro culture and molecular and morphological changes induced by insulin and BMP-4. Front. Vet. Sci. 9, 1068602; doi: 10.3389/fvets.2022.1068602. Ghazi, A.M., Al-Bayati, A.M.A. and Janabi, A.H.D. 2024. Metabolomics-detected alterations generated by phytosomal propolis and phytosomal lycopene in male rats with induced benign prostatic hyperplasia. Iraqi J. Vet. Sci. 38(Supplement I-IV), 7–15; doi: 10.33899/ijvs.2024.147764.3531. Gholami, S., Behrestaghi, L.E., Sarvi, S., Alizadeh, A. and Spotin, A. 2021. First description of Echinococcus ortleppi (G5 genotype) in Iran. Parasitol. Int. 83, 102316; doi: 10.1016/j.parint.2021.102316. Guo, B., Zhao, L., Zhao, L., Mi, R., Zhang, X., Wang, B., Guo, G., Ren, Y., Qi, W. and Zhang Z. 2023. Survey and molecular characterization of Echinococcus granulosus sensu lato in Xinjiang, China. Pathogens. 12(1), 134; doi: 10.3390/pathogens12010134. Hodžić, A., Alić, A., Spahić, A., Harl, J. and Beck, R. 2022. Genetic diversity of Echinococcus granulosus sensu lato from animals and humans in Bosnia and Herzegovina. Parasites Vectors 15(1), 457. Hogea, M.O., Ciomaga, B.F., Muntean, M.M., Muntean, A.A., Popa, M.I. and Popa, G.L. 2024. Cystic echinococcosis in the early 2020s: a review. Trop. Med. Infect. Dis. 9(2), 36; doi: 10.3390/tropicalmed9020036. Janabi, A.H.D. 2021. Molecular docking analysis of anti-severe acute respiratory syndrome-coronavirus 2 ligands against spike glycoprotein and the 3-chymotrypsin-like protease. J. Med. Signals Sens. 11(1), 31–36; doi: 10.4103/jmss. JMSS_25_20. Jesudoss Chelladurai, J.R.J., Quintana, T.A., Johnson, W.L., Schmidt, C., Righter, D. and Howey, E. 2024. Cystic echinococcosis in cattle and sheep in the USA. Parasit. Vectors. 17(1), 128; doi: 10.1186/s13071-024-06192-x. Karim, S., Mansour, K., Janabi, A.H.D. and Al-Nakeeb, N. 2019. First phylogenetic characterization of Pseudocowpox virus from cattle in Al-Qadisiyah province, Iraq. Iraqi J. Vet. Sci. 33(1), 123–126; doi: 10.33899/ijvs.2019.125525.1047. Kheninef, A., Celik, F., Aissaoui, L. and Simsek, S. 2024. Molecular characterization of hydatid cyst isolates from Algeria. Parasitol. Res. 123(3), 159; doi: 10.1007/s00436-024-08176-3. Liguori, G., Costagliola, A., Lombardi, R., Paciello, O. and Giordano, A. 2023. Human-animal interaction in animal-assisted interventions (AAIs): zoonosis risks, benefits, and future directions—A one health approach. Animals. 13(10), 1592; doi: 10.3390/ani13101592. Oliveira, F.M.S., Cruz, R.E., Pinheiro, G.R.G. and Caliari, M.V. 2022. Comorbidities involving parasitic diseases: a look at the benefits and complications. Exp. Biol. Med. 247(20), 1819-1826; doi: 10.1177/15353702221108387. Omondi, H.A., Gitau, G., Gathura, P., Mulinge, E., Zeyhle, E., Kimeli, P. and Bett, B. 2020. Prevalence and genotyping of Echinococcus granulosus in Kenya. J. Helminthol. 94, e205; doi: 10.1017/S0022149X20000899. Pereira, I., Hidalgo, C., Stoore, C., Baquedano, M.S., Cabezas, C., Bastías, M., Riveros, A., Meneses, C., Cancela, M., Ferreira, H.B., Sáenz, L. and Paredes, R. 2022. Transcriptome analysis of Echinococcus granulosus sensu stricto protoscoleces. Vet. Res. 53(1), 8; doi: 10.1186/s13567-022-01022-3. Shahabi, S., Sarkari, B. and Barazesh, A. 2021. Echinococcus granulosus sensu stricto G1 is the predominant genotype in human and livestock isolates from Turkey and Iran, based on mitochondrial nad 5 gene differentiation. Parasites Vectors 14(1), 369. Vishniakov, D., Turdumambetova, M., Matkerimova, N., Dzhusupov, K., Sultanbaeva, Z. and Rafibekov, E. 2024. Neglected yet pervasive: echinococcosis awareness and prevention capacity in Kyrgyzstan. Parasitology 151(13), 1487–1495; doi: 10.1017/S0031182024001343. Widdicombe, J., Basáñez, M.G., Entezami, M., Jackson, D., Larrieu, E. and Prada, J.M. 2022. The economic evaluation of cystic echinococcosis control strategies focused on zoonotic hosts: a scoping review. PLoS Negl. Trop. Dis. 16(7), e0010568; doi: 10.1371/journal.pntd.0010568. Xie, Y., Shi, D., Wang, X., Guan, Y., Wu, W. and Wang, Y. 2023. Prevalence trend and burden of neglected parasitic diseases in China from 1990 to 2019: findings from the global burden of disease study. Front. Public Health. 11, 1077723; https://doi.org/10.3389/fpubh.2023.1077723. | ||
| How to Cite this Article |
| Pubmed Style Resen DS, Jarad NI. Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq. Open Vet. J.. 2025; 15(7): 2948-2958. doi:10.5455/OVJ.2025.v15.i7.6 Web Style Resen DS, Jarad NI. Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq. https://www.openveterinaryjournal.com/?mno=243136 [Access: January 24, 2026]. doi:10.5455/OVJ.2025.v15.i7.6 AMA (American Medical Association) Style Resen DS, Jarad NI. Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq. Open Vet. J.. 2025; 15(7): 2948-2958. doi:10.5455/OVJ.2025.v15.i7.6 Vancouver/ICMJE Style Resen DS, Jarad NI. Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq. Open Vet. J.. (2025), [cited January 24, 2026]; 15(7): 2948-2958. doi:10.5455/OVJ.2025.v15.i7.6 Harvard Style Resen, D. S. & Jarad, . N. I. (2025) Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq. Open Vet. J., 15 (7), 2948-2958. doi:10.5455/OVJ.2025.v15.i7.6 Turabian Style Resen, Doaa Sabri, and Noor Idan Jarad. 2025. Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq. Open Veterinary Journal, 15 (7), 2948-2958. doi:10.5455/OVJ.2025.v15.i7.6 Chicago Style Resen, Doaa Sabri, and Noor Idan Jarad. "Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq." Open Veterinary Journal 15 (2025), 2948-2958. doi:10.5455/OVJ.2025.v15.i7.6 MLA (The Modern Language Association) Style Resen, Doaa Sabri, and Noor Idan Jarad. "Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq." Open Veterinary Journal 15.7 (2025), 2948-2958. Print. doi:10.5455/OVJ.2025.v15.i7.6 APA (American Psychological Association) Style Resen, D. S. & Jarad, . N. I. (2025) Genotyping and phylogenetic analysis of Echinococcus granulosus isolated from human, sheep, and cattle samples in Iraq. Open Veterinary Journal, 15 (7), 2948-2958. doi:10.5455/OVJ.2025.v15.i7.6 |