| Research Article | ||
Open Vet. J.. 2025; 15(9): 4219-4234
Open Veterinary Journal, (2025), Vol. 15(9): 4219-4234 Research Article One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humansNourhan Eissa1, Marwa B. Salman2, Abdelgayed M. Younes3, EL Shymaa A. Mohamed4, Ashraf M. Abu-Seida5*, Abdulrahman Abdulkarim6 and Asmaa I. Zin Eldin71Department of Animal Hygiene and Zoonoses, Faculty of Veterinary Medicine, University of Sadat City, Cairo, Egypt 2Department of Zoonotic Diseases, National Research Center, Giza, Egypt 3Hydrobiology Department, National Research Center, Giza, Egypt 4Department of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt 5Department of Surgery, Anesthesiology and Radiology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt 6Faculty of Veterinary Medicine, Omar Almukhtar University, Bayda, Libya 7Department of Microbiology and Immunology, National Research Center, Giza, Egypt *Corresponding Author: Ashraf M. Abu-Seida. Department of Surgery, Anesthesiology, and Radiology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt. Email: ashrafseida [at] cu.edu.eg Submitted: 08/06/2025 Revised: 30/07/2025 Accepted: 06/08/2025 Published: 30/09/2025 © 2025 Open Veterinary Journal
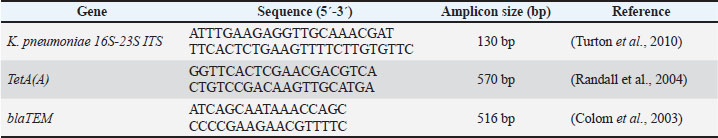
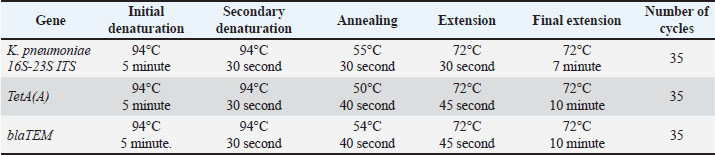
AbstractBackground: Worldwide, zoonotic diseases represent serious risks to public health, underscoring the need for efficient surveillance techniques. The One Health concept has gained popularity as a comprehensive paradigm for tackling zoonotic disease surveillance because it acknowledges the correlation of pathogens, animals, humans, and the surrounding environment. Public health is now concerned about antibiotic-resistant Klebsiella pneumoniae (K. pneumoniae), which causes severe impurities in animals and humans, leading to morbidity and mortality. Aim: This study examined the collaborative efforts between human and animal (cattle and horses) health sectors, as well as other pertinent risk factors, in implementing the One Health surveillance strategy against klebsiellosis in one Egyptian district. In addition, this study analyzed some K. pneumoniae antimicrobial resistance genes to improve future medical treatment strategies against zoonotic klebsiellosis. Methods: Klebsiella pneumoniae was isolated from 50 cattle raw milk samples, 50 horse nasal swabs, and 50 human sputum samples using MacConkey’s agar and Xylose lysine deoxycholate media with a detailed explanation of the associated risk factors of infection acquisition. Then, antibiotic sensitivity tests for the positive samples were conducted on plates of Mueller–Hinton agar. In addition, polymerase chain reaction (PCR) was performed on the positive isolates to detect the pathogen and antibiotic resistance genes. Results: Klebsiella pneumoniae was effectively isolated from 8% of milk samples, 12% of nasal swabs, and 16% of sputum samples. Upon conducting antibiotic sensitivity tests, the isolated organisms showed resistance to 100% amoxicillin-clavulanic, 93.3% cefotaxime, 86.66% cepaxime, and 66.66% tetracycline. Only 11 of 18 positive K. pneumoniae samples were PCR-positive. Only 10 of 11 positive PCR samples were positive for the TetA(A) gene; however, all 11 samples were positive for the blaTEM gene. Conclusions: By focusing on various risk factors of disease acquisition, this study sheds light on the zoonotic hazard of K. pneumoniae that has been hitherto underestimated. It also emphasizes the significance of integrating One Health into surveillance for antibiotic-resistant klebsiellosis and the necessity of ongoing innovation and collaboration to improve the efficacy of upcoming surveillance plans in Egypt to control the spread of the disease between animals and humans. Keywords: Cattle, Horses, Humans, K. pneumoniae, Surveillance, One Health. IntroductionWorldwide, the term “zoonosis” refers to any illness that humans could contract from domestic or even wild animals (Zin Eldin et al., 2023; Eissa, 2024a, b; Zain Eldeen et al., 2024; Ramzy et al., 2025). Approximately 75% of recently discovered infectious diseases in humans have zoonotic origins, meaning they mostly stem from animals (Bedair et al., 2024; Elsobky et al., 2024; Salman et al., 2024a; Zin Eldin et al., 2025). The Gram-negative zoonotic Klebsiella pneumoniae is an opportunistic bacterium that can cause various illnesses in humans and animals (Hu et al., 2021). The quality of milk and milk products in cattle is negatively impacted by a prevalent environmental factor that causes clinical and subclinical bovine mastitis in cattle (Huang et al., 2020; Zin Eldin et al., 2025). Because K. pneumoniae does not react well with antibiotic therapy, it can induce toxic shock and animal mortality, making its mastitis more severe (Sugiyama et al., 2022). In horses, the majority of clinical signs appear in the respiratory and urinary systems, where the annual frequency of K. pneumoniae multidrug-resistant (MDR) strains ranged from 11.7% to 51.5% from 2006 to 2019 (Léon et al., 2020). Furthermore, K. pneumoniae has been linked to sepsis, infertility problems, metritis, and miscarriage in mares (Akter et al., 2021). On the other hand, human klebsiellosis is the third leading cause of healthcare illnesses, including urinary tract infections, ventilator-associated pneumonia, bacteremia, and a serious community-acquired infection that manifests as fasciitis, meningitis, and liver abscesses (Holt et al., 2015). Klebsiella pneumoniae is the main causative agent of nosocomial pneumonia, which is more common in older people or immunocompromised individuals (Magill et al., 2018). Owing to the rise in the prevalence of microbial diseases over the past few decades, antimicrobial medications have been used continuously and uncontrollably for both prevention and treatment in several countries worldwide. Consequently, many strains of bacteria developed specialized and MDR (Salman et al., 2024a, b; Tanwar et al., 2014; Zain Eldeen et al., 2024). Since the public health panic of the late 20th century was sparked by the MDR Salmonella typhimurium DT104 clone, antibiotic-resistant bacteria (AMR) have long existed (Dunn et al., 2019; Zin Eldin et al., 2025). Gram-negative bacteria have recently developed a variety of resistance mechanisms against the in-use antimicrobials (Munoz-Price and Quinn, 2009; Eissa and Harb, 2023; Salman et al., 2024b; Zin Eldin et al., 2025). Since the 1970s, commonly shared plasmids among bacteria such as Escherichia coli, K. pneumoniae, Klebsiella oxytoca, Salmonella sp., and Enterobacter sp. have been linked to the transmission of MDR isolates in clinical settings (Toledano-Tableros et al., 2021). According to a 2019 Centers for Disease Control and Prevention report, AMR against Gram-negative bacteria caused around 4.95 million fatalities worldwide, whereas in the United States, over 2.8 million AMR infections occur annually, leading to over 35,000 deaths (CDC, 2019). Furthermore, the World Health Organization (WHO) prioritizes K. pneumoniae as a member of the six members of the ESKAPE group because of its capacity to “escape” the effects of antibiotics (WHO, 2017). In Egypt, K. pneumoniae, a carbapenem-resistant Enterobacteriales, is a significant pathogen in nosocomial infections (Abdel-Wahab et al., 2013). Nonetheless, there are few data on the representation of virulence markers and their association with clinical origin and MDR. To deploy One Health surveillance tactics against klebsiellosis in Egypt, the current study attempted to shed light on several antimicrobial resistance genes of isolated K. pneumoniae strains from animals and humans with the associated epidemiological risk factors of the disease acquisition and shed light on the importance of cooperative efforts between the human and animal health sectors (cattle and horses), communication, and data sharing between these sectors to improve zoonotic klebsiellosis early identification, quick reaction, and efficient control. Materials and MethodsCollection and processing of samplesThe present zoonotic study was conducted on two fronts: first aspect, the animal side, 50 raw cows’ milk samples (30 ml each, were put in sterile plastic containers following teat wiping with 70% ethanol and neglecting the first drops of foremilk) were collected from seemingly healthy cattle of different ages of the native breed as previously mentioned by Byomi et al. (2018, 2019b), Salman et al. (2024a), and Zin Eldin et al. (2025). In addition, 50 horse nasal swabs were obtained (sterile cotton swabs placed in peptone water broth; Microxpress Pvt. Ltd., India) from the nasal cavity of each horse, and the swabs were reinserted into sterile, labeled tubes containing 5 ml of peptone water broth (Bekele et al., 2024). As previously mentioned by Bekele et al. (2024), samples were taken from draft horses that displayed respiratory symptoms such as fever, nasal discharges, coughing, sneezing, and enlarged lymph nodes. The horses were of various ages. Regarding the second aspect, a total of 50 human sputum samples (in sterile swabs) of different ages, sexes, and occupations, with close contact with the aforementioned animal samples/species, were obtained for the human side of the study. According to Myatt (2017), the studied humans displayed different signs of respiratory manifestations (as previously mentioned in horses), which are resistant to various known common antibiotics. Between December 2022 and November 2023, all human and animal samples were taken from a teaching farm in Menoufia governorate, Egypt, along with a history questionnaire specific to the species under investigation. The gathered samples were promptly sent in an icebox (2°C–5°C) to the Department of Zoonoses, Faculty of Veterinary Medicine, University of Sadat City, Menoufia Governorate, Egypt, where they were immediately examined. Isolation of K. pneumoniaeMacConky’s agar and Xylose lysine deoxycholate (XLD) plates (100 μl each) (Oxoid, Basingstoke, UK) were equally spread with the appropriate dilutions of the collected samples. The cultured and control plates were then incubated overnight (16–18 hours) at 37°C. Following incubation, the putative Klebsiella colonies were chosen and preserved in pure form to facilitate additional identification by different biochemical tests and polymerase chain reaction (PCR). Antimicrobial sensitivity assayThe following six widely used antibiotic discs (WHO designated these classes of antibiotics as a “critical concern”), were used to screen the isolated organisms for antibiotic sensitivity tests, in agreement with the values of the Clinical & Laboratory Standards Institute (CLSI, 2018): imipenem (IPM10), which belongs to the carbapenems group; amoxicillin-clavulanic (AMC30) which is β-lactam (Penicillin) and β-lactam inhibitor antibiotics, Ceftriaxone (CRO30), which is a cefalosporin 3rd generation; cefotaxime (CTX30), which is a cefalosporin 3rd generation; cephalothin (KF30), which is cephalosporin 1st generation; and tetracycline (TE 10). Sterile cotton swabs were added to the broth, squeezed, and spread onto Mueller–Hinton agar plates. Nutrient broth tubes grown with the resulting Klebsiella isolates were visually matched with standard McFarland tubes (0.5 McFarland in accordance with the Clinical Laboratory Standards Institute guidelines). Using sterile forceps, antibacterial discs were aseptically placed on the surface of the plate at the proper spacing, and they were then incubated aerobically for 24 hours at 37°C. Measure the diameter of the zone of inhibition (if any) for every antibiotic used using a metric ruler. The susceptibility of the resultant bacteria was ascertained by comparing the measurements obtained for each used particular antibiotic to the standards table (Wheat, 2001). Molecular detection of K. pneumoniae using conventional PCRTo detect K. pneumoniae (16S-23S ITS gene), DNA was first extracted from positive cultured samples in accordance with the QIAamp DNA mini kit instructions (cat. no. 51304, Qiagen, Germany, GmbH) according to Gomaa (2021), followed by screening all of the obtained positive PCR isolates for β-lactam (blaTEM gene), the most frequently observed ESBL gene as a result of mutations in the β-lactamase gene (Seyedjavadi et al., 2016), and tetracycline (TetA(A) gene) resistance. PCR reactions were performed using a thermocycler (Biometra T3 thermocycler, Göttingen, Germany), with a 25 µl reaction volume. Finally, the extracted DNA was electrophoresed on a 1% agarose gel. Gel visualization and analysis were performed using a gel documentation system (Alpha-Innotech, USA). Tables 1 and 2 present the primers used and the thermal cycling conditions, respectively. Table 1. PCR genes, primers, and expected product sizes of bands used in the current study.
Table 2. Cycling conditions (adjusted by thermal cycler) used for gene detection.
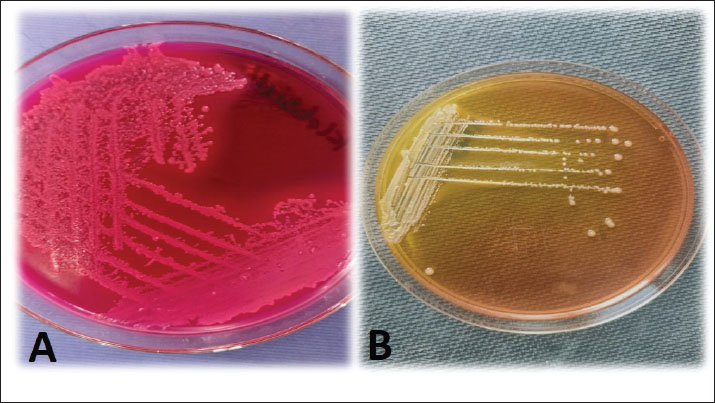
Statistical analysisVersion 17 of the Statistical Package for the Social Sciences was utilized to analyze all the data using the chi-square test (where a p-value of >0.05 was statistically insignificant, a p-value ≤ 0.05 was statistically significant, and <0.01 was highly significant), as mentioned by Byomi et al. (2019a, b) and Zin Eldin et al. (2025). Additionally, percentages were calculated to characterize the infection frequency. Ethical approvalThe Institutional Animal Care and Use Committee granted consent for this study under the direction of the University of Sadat City’s Faculty of Veterinary Medicine (Ethical approval number: VUSC-037-1-23). ResultsKlebsiella pneumoniae isolation on MacConky’s agar and XLD platesKlebsiella pneumoniae isolates showed pink mucoid colonies on MacConkey’s agar plate and yellow mucoid colonies on XLD plate (Fig. 1). Of the 150 samples examined, 18 isolates were identified as K. pneumoniae with a total occurrence of 12%. The highest occurrence was 16% (8/50) in human samples, followed by 12% (6/50) in horse nasal swabs and 8% (4/50) in cattle milk.
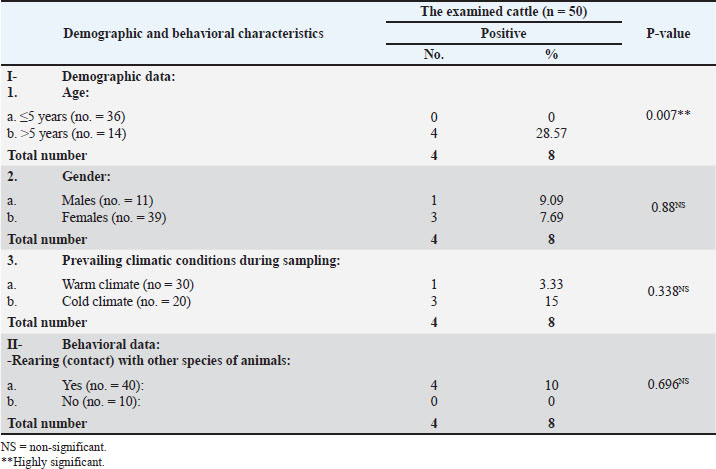
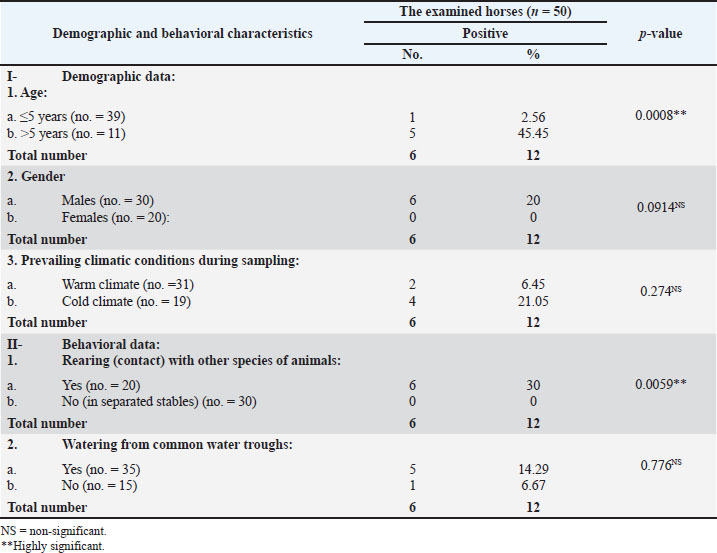
Fig. 1. A representative K. pneumoniae isolates showing mucoid pink colonies on MacConkey’s agar plate (A) and mucoid yellow colonies on XLD plate (B). The obtained biochemical findings showed that K. pneumoniae was non-motile and negative for indole synthesis, but showed positive reactions for lactose fermentation, citrate, catalase, and urease consumption. These observations distinguished K. pneumoniae from other germs. Occurrence of K. pneumoniae in relation to the demographic risk factors of the different species examinedTables 3 and 4 show that the infection occurrence among animals in the age group ˃5 years to be 28.57% (4/14) and 45.45% (5/11), 9.09% (1/11), and 20% (6/30) among males, 15% (3/20) and 21.05% (4/19) during cold climate, and 10% (4/40) and 15% (6/40) among animals in close contact with other animal species among the examined cattle and horses, respectively. Furthermore, the occurrence of K. pneumoniae infection increased by 14.29% (5/35) among horses with a habit of watering from common water troughs. Table 3. The occurrence of K. pneumoniae bacteria in relation to the demographic and behavioral data of the cattle examined (n=50).
Table 4. The occurrence of K. pneumoniae bacteria in relation to the demographic and behavioral data of the examined horses (n=50)
Table 5 presents the occurrence of K. pneumoniae to be increased among human individuals of age group (˃40–60) years to be 50% (5/10), males 17.5% (7/40), during cold climate 24% (6/25), butchers 30% (3/10), individuals in close contact with animal and bird species 16.67% (7/42), consumers of raw milk and raw milk products 50% (6/12), smokers 33.33% (5/15), immunocompromised humans suffering from hepatitis C virus 30% (6/20), diabetes mellitus 30% (3/10), and even those suffering from heart disorders 20% (1/5). Table 5. The occurrence of K. pneumoniae bacteria in relation to demographic data, behavioral risk factors, and immunological (health) status of the examined individuals (n=50).
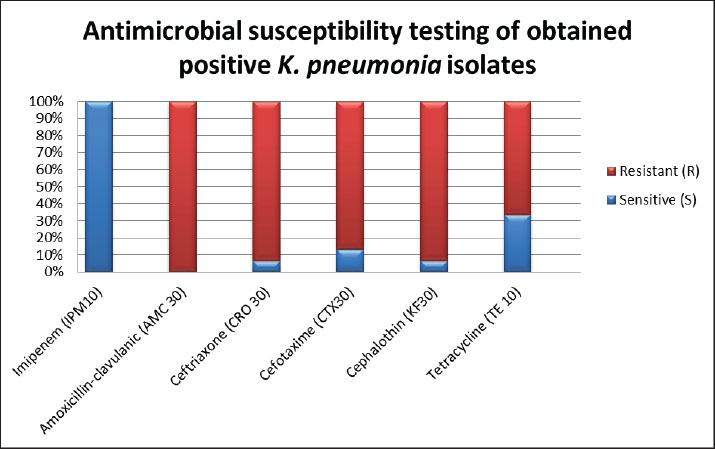
Antimicrobial susceptibility testing resultsAll positive isolates were tolerant to amoxicillin-clavulanic and ceftriaxone (CRO30), and 93.3% of the samples showed resistance to cephalothin (KF30), as shown in Figures 2 and 3. In addition, 86.66% of the samples were tolerant of cefotaxime (CTX 30). All samples reacted sensitively to imipenem (IPM10), whereas 66.6% of samples showed resistance to tetracycline (TE 10).
Fig. 2. Antimicrobial susceptibility testing of positive K. pneumoniae isolates.
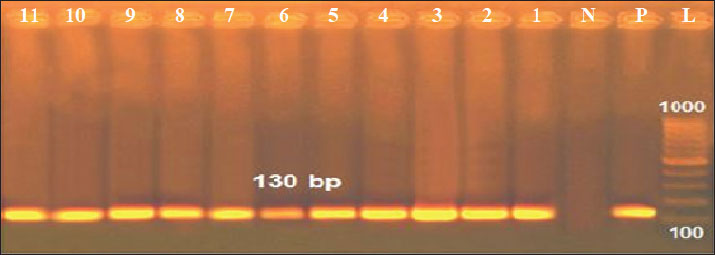
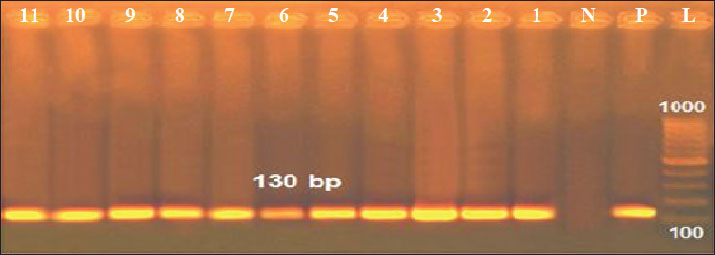
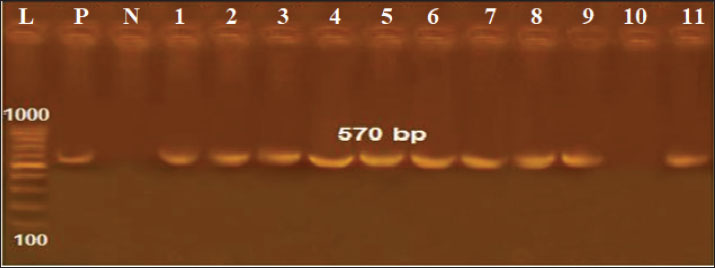
Fig. 3. Antimicrobial susceptibility testing of positive K. pneumoniae isolates on Mueller-hinton agar plates. Results of the PCROf the 18 positive cultured K. pneumoniae, only 11 isolates were positive by conventional PCR targeting the K. pneumoniae 16S-23S ITS gene (61.11%), as shown in Figure 3. Out of these 11 isolates, 2 of 4 were positive in raw cattle milk (50%), 3 of 6 were positive in horse nasal swabs (50%), and 6 of 8 were positive in human sputum samples (75%). Furthermore, by applying PCR targeting TetA(A) and blaTEM genes on the 11 positive PCR samples, all samples were positive for the blaTEM gene (516 bp), whereas only 10 samples were positive for the TetA(A) gene (570 bp), as shown in Figures 4–6.
Fig. 4. Agarose gel electrophoresis of the PCR product amplified from the K. pneumoniae 16S-23S ITS gene (130 bp). Lane L: 100 bp DNA ladder; Lane P: positive control; Lane N: negative control; Lanes 1–11: Positive samples.
Fig. 5. Agarose gel electrophoresis of the PCR product amplified from the blaTEM gene (516 bp). Lane L: 100 bp DNA ladder; Lane P: positive control; Lane N: negative control; Lanes 1–11: Positive samples.
Fig. 6. Agarose gel electrophoresis of the PCR product amplified from the TetA(A) gene (570 bp). Lane L: 100 bp DNA ladder; Lane P: positive control; Lane N: negative control; Lanes 1–11: Positive samples. DiscussionEgypt’s national antimicrobials sustainable strategy is not rigorously implemented by regulations despite being established (El-Sokkary et al., 2021; Nasr and Eissa, 2025a). Sobur et al. (2019) and Ramzy et al. (2025) reported that Egyptian veterinarians are still using different antimicrobials to treat and prevent zoonotic illnesses and aid in animal growth. The present results showed that 16%, 12%, and 8% of the isolates were found in human sputum, horse nasal swabs, and raw cattle milk, respectively. The total percentage of isolated K. pneumoniae from all examined samples was 12%. The results showed that 8% of the isolates were found in the milk of raw cattle. These findings are relatively consistent with those of Kaczorek-Lukowska et al. (2021) and Shehata et al. (2024), who reported an isolation percentage of 9.87% in Poland and 6.4% in the Alexandria governorate, Egypt. However, higher percentages of K. pneumoniae were presented in Egypt as: 41% in ElGharbia governorate (Bedawy et al., 2024), Behaira governorate (22.7%) (Alaa et al., 2024), and Cairo governorate (14.6%) (Abebe et al., 2016). Moreover, a higher K. pneumoniae occurrence rate of 45.29% was found in India (Koovapra et al., 2016) and 40% in Iran (Enferad and Mahdavi, 2020). A comparable 12.12% K. pneumoniae isolation rate was found in horses in a Turkish study (Borum, 2022). In contrast, a lower isolation rate of 3.3% was previously reported in Australia (Loncaric et al., 2020). In Egypt, Osman et al. (2021) reported a higher K. pneumoniae isolation rate of 26.3% in the Cairo governorate, whereas Arafa et al. (2022) observed a lower isolation rate of 6.28% in the Giza governorate. The human outcome was consistent with the occurrence rates of Egyptian observations, which were 14% in the governorates of Assiut, El-Minia, and Sohag (Agmy et al., 2013) and 14.9% in the governorate of Cairo (Sobhy et al., 2015). Conversely, higher percentages of K. pneumoniae have been reported in India (Shilpa et al., 2016), the Sharkia governorate, Egypt (Al-Baz et al., 2022), and China (Liu et al., 2023). Variations in reported results between the current study and different published investigations would be referred to the variations in techniques used to conduct the investigations, and differences in the examined species, geographical status, prevalent climatic conditions, demographic, behavioral, and immunological status of the tested species (Eissa, 2024c). Veterinarians now play a more involved role than simply waiting for a sickness to manifest before prescribing medication (Singh, 2010; Eissa, 2024c; Zin Eldin et al., 2025). Therefore, to assist physicians and epidemiologists in the future with suspicion, differential diagnosis, or even ruling out the occurrence of the pathogen in various communities, demographic and behavioral data were recorded among the species under examination. Older age (˃5 years), male sex, cold environment, interaction with other animal species (mixed breeding), and drinking from communal water troughs were the demographic risk variables linked to increased K. pneumoniae infection among the examined cattle and horses. Regarding the animal’s age, the obtained results were consistent with those of Adler et al. (2015), who did not find any correlation between klebsiellosis and advanced age. Our results showed that mature animals (˃5 years) were exposed to higher levels of antibiotics as prescriptions and feed additives, as well as higher levels of infection sources (soil, water, urine, aerosols, dung, etc.) (Ribeiro et al., 2022). Regarding gender, Shnaiderman-Torban et al. (2020) found that males (stallions and bulls) were more prone to klebsiellosis, which is likely related to hormonal influences. The current results revealed that klebsiellosis occurred during the cold climate. This is consistent with the results of Ribeiro et al. (2022), Eissa and Nasr (2025), El-Khadry et al. (2025), and Nasr and Eissa (2025a, b). This is because aerosolization between animals raised in close contact may facilitate pathogen transmission during this time, and cold temperatures may weaken the immune responses of animals, causing cold stress and enhancing the airborne transmission of the pathogen (Ribeiro et al., 2022). Conversely, Gomaa (2021) and Shehata et al. (2024) observed increased K. pneumoniae infection rates in hot temperatures. The risk of acquiring klebsiellosis from mixed breeding of different animal species could be explained by K. pneumoniae being a major cause of clinical mastitis in cows and a pathogen of mammary infections in cattle for many years. The organism is extensively distributed in the environment of dairy farms, including manure, feces, water, and various animal dwellings (sand, sawdust, and straw) (Schukken et al., 2012; Nasr and Eissa, 2025a, b). This fact would also explain why infections among cattle and horses raised together are more common, as evidenced by the findings that horses regularly consuming water from communal troughs are more likely to be infected than horses kept in separate stables (Kumar and Kumari, 2013). This is because K. pneumoniae can persist in a broad range of reservoirs, including livestock, water, and untreated wastewater, biting midges and houseflies inside animal stables, and closed crowded houses (Mohammed et al., 2016). Regarding klebsiellosis, K. pneumoniae infection increased in the age group of ˃40–60 years, which was corroborated by Meatherall et al. (2009) and Chiu et al. (2011). This observation could be explained by the fact that the capacity of neutrophils to phagocytize virulent K1/K2 K. pneumoniae declines with age (Chiu et al., 2011). Furthermore, this age group (˃40–60 years) had the longest exposure duration to various sources of infection, meaning that they had greater contact with contaminated farm animals and soil during various occupation activities. In contrast, the age group of ˃60 years comprised older adults retirees and unemployed people (Byomi et al., 2019a). The observed increased occurrence of K. pneumoniae infection among men may be explained by several factors, which were also confirmed by the conclusions of Chakraborty et al. (2016) and Rao et al. (2021). First, there are more men than women working on farms and clinics, managing animals, and performing obstetrics. Second, more men than women work in slaughterhouses. Third, studies conducted by Chang et al. (2017) and Eissa (2024c) revealed that women are protected against the majority of infections by the female sex hormone, 17-beta-estradiol. The results showed that infection was higher in colder climates, which concurs with the conclusions of Brat et al. (2016), who were able to isolate several anthropogenic bacteria, including Klebsiella, under harsh winter conditions in the Antarctic. However, in warmer climates, studies by Schwab et al. (2014) and Chang et al. (2017) reported higher K. pneumoniae infection rates. According to Klemmer et al. (2018) and Byomi et al. (2019a), a correlation may exist between the slightly higher positivity for K. pneumoniae infection among the examined animal species (cattle and horses) in cold climates and the slightly higher prevalence of the infection among the examined humans who had close contact with those animals. According to the current investigation, butchers had the highest rate of K. pneumoniae positivity among all other groups. This finding may be explained by the fact that butchers are the most frequently examined human group and have the greatest contact with animal carcasses, body fluids, and blood. Consequently, they are the most frequently exposed to various sources of infection (Byomi et al., 2019a; Deepan et al., 2023). The behavioral data collected from the examined human subjects corroborated this observation, showing a higher percentage of K. pneumoniae infection in those who had frequent contact with other animal and bird species as well as in those who regularly consumed raw milk and raw milk products (Elrefaey and Eissa, 2025). Furthermore, tobacco users were more vulnerable to contracting K. pneumoniae infection than non-smokers (Aljanaby et al., 2018). According to Byomi et al. (2019a), smoking induces respiratory distress because it has detrimental mechanical impacts on the respiratory mucosa as well as detrimental immunomodulation, which facilitates the invasion of respiratory macrophages by pneumonic anthropogenes. Finally, by examining the relationship between individuals’ immunological status and their susceptibility to K. pneumoniae infection, it was found that those with heart problems, diabetes mellitus, and the hepatitis C virus were more susceptible to klebsiellosis, which is consistent with the findings of Chakraborty et al. (2016), Mohammed et al. (2016), and Aljanaby et al. (2018). The current study also used Mueller–Hinton agar plates to conduct the antibiogram susceptibility analysis of positive K. pneumoniae isolates from various species under investigation. Each of the obtained positive K. pneumoniae isolates was tolerant to the following antibiotics: 100% amoxicillin-clavulanic, 100% ceftriaxone (CRO30), 93.3% cephalothin (KF30), 86.66% cefotaxime (CTX30), and 66.6% Tetracycline (TE10). Based on published data on K. pneumoniae antibiogram testing, resistance against cefotaxime was reported by Namratha et al. (2015), resistance against amoxicillin was demonstrated by Chakraborty et al. (2016), resistance against amoxicillin, cefotaxime, and tetracycline was reported by Aljanaby et al. (2018), and resistance against amoxicillin-clavulanic and tetracycline was reported by Shehata et al. (2024). Furthermore, it should come as no surprise that only 11 isolates (61.11%) out of the 18 positive cultured samples tested positive by conventional PCR targeting the K. pneumoniae 16S–23S ITS gene. This is because, as previous studies have shown, PCR is time-dependent and loses value over time (Lucchese et al., 2015; Byomi et al., 2019a). The samples were cultured immediately, and their DNA was stored as aliquots at −20°C until PCR testing. Furthermore, samples of raw milk from calves, horse nasal swaps, and human sputum had PCR-positive rates of 50%, 50%, and 75%, respectively. Following further PCR targeting the TetA(A) and blaTEM genes, 90.91% of the isolates under examination tested positive for the TetA(A) gene (10/11) and 100% tested positive for the blaTEM gene (11/11). Selmi et al. (2022), Al-Sheboul et al. (2023), Dan et al. (2023), and Paniagua-Contreras et al. (2023) obtained similar results. The advent of highly pathogenic and MDR strains of K. pneumoniae that have been isolated from humans and several animal species has increased the number of fatalities. Furthermore, scant research has been conducted on the molecular characterization of genes associated with infection type and resistance to antibiotics in K. pneumoniae strains isolated from various species (Paniagua-Contreras et al., 2023). The majority of research has focused on other Enterobacteriaceae members such as E. coli, Salmonella, and Campylobacter spp. (Elsobky et al., 2025), and there is a dearth of Egyptian research on zoonotic MDR K. pneumoniae in various species with associated risk factors of disease acquisition. This may be because the zoonotic hazard of the disease is still underestimated. Therefore, the current work is noteworthy and may contribute to the future advancement of Egyptian medical treatment protocols for MDR K. pneumoniae illnesses in humans and animals. Furthermore, this work will serve as a solid foundation for any upcoming epidemiological surveys, mapping, and comparisons of the many risk variables associated with klebsiellosis in humans, horses, and cattle. ConclusionThis study highlights the risk posed by zoonotic opportunistic K. pneumoniae. This study also focuses on the significant rise in the emergence of AMR-K. pneumoniae in both animal and human populations in Egypt, highlighting the importance of One Health in the context of the surveillance of AMR-K. pneumoniae and the ongoing need for collaboration and innovation to enhance the effectiveness of upcoming surveillance initiatives and future preventive measures in Egypt. Moreover, the present research declared that no obtained strain was resistant to imipenem, which is a positive and noteworthy result, emphasizing that the current findings may contribute to ongoing monitoring efforts and help in the prevention of CRE emergence. AcknowledgmentsNot applicable. Conflict of interestThere are no conflicts of interest. FundingNot applicable. Authors’ contributionsNE, MBS, and AIZE contributed to the study design, sampling process, laboratory work, scientific writing process, and data analysis. AMY and EAM also collaborated in sampling and data processing. AMA and AA assisted with drafting and efficient revision of the manuscript. All authors have read and approved the final manuscript. Data availabilityThe manuscript included all of the relevant data. ReferencesAbdel-Wahab, F., Ghoneim, M., Khashaba, M., El-Gilany, A.H. and Abdel-Hady, D. 2013. Nosocomial infection surveillance in an Egyptian neonatal intensive care unit. J. Hosp. Infect. 83(3), 196–199; doi:10.1016/j.jhin.2012.10.017 Abebe, R., Hatiya, H., Abera, M., Megersa, B. and Asmare, K. 2016. Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet. Res. 12, 1–11; doi:10.1186/s12917-016-0905-3 Adler, A., Sturlesi, N., Fallach, N., Zilberman-Barzilai, D., Hussein, O., Blum, S.E., Klement, E., Schwaber, M.J. and Carmeli, Y. 2015. Prevalence, risk factors, and transmission dynamics of extended-spectrum-β-lactamase-producing Enterobacteriaceae: a national survey of cattle farms in Israel in 2013. J. Clin. Microbiol. 53(11), 3515–3521; doi:10.1128/jcm.01915-15 Agmy, G., Mohamed, S., Gad, Y., Farghally, E., Mohammedin, H. and Rashed, H. 2013. Bacterial profile, antibiotic sensitivity and resistance of lower respiratory tract infections in upper Egypt. Medit. J. Hematology Infect. Dis. 5, e2013056; doi:10.4084/MJHID.2013.056 Akter, R., El-Hage, C.M., Sansom, F.M., Carrick, J., Devlin, J.M. and Legione, A.R. 2021. Metagenomic investigation of potential abortigenic pathogens in fetal tissues from Australian horses. BMC Genomics 22, 1–14; doi:10.1186/s12864-021-08010-5 Alaa, O.A., Elzeftawy, H.M. and Salama, H.F. 2024. Advanced studies on extended spectrum beta-lactamase producing Enterobacteriaceae in dairy cattle farms at Behaira province. J. Adv. Vet. Res. 14(3), 470–474; doi:10.1016/j.javi.2018.09.010 Al-Baz, A.A., Maarouf, A., Marei, A. and Abdallah, A.L. 2022. Prevalence and antibiotic resistance profiles of carbapenem-resistant Klebsiella pneumoniae isolated from Tertiary Care Hospital, Egypt. Egypt. J. Hosp. Med. 88(1), 2883–2890; doi:10.21608/ejhm.2022.242765 Aljanaby, A.A.J., Tuwaij, N.S. and Al-khilkhali, H.J. 2018. Antimicrobial susceptibility patterns of Klebsiella pneumoniae isolated from older smokers and non-smokers of inpatients in intensive care unit infected with chronic pneumonia in AL-Najaf hospital, Iraq. J. Pharmaceut. Sci. Res. 10(5), 1093–1097. Al-Sheboul, S.A., Al-Madi, G.S., Brown, B. and Hayajneh, W.A. 2023. Prevalence of extended-spectrum β-lactamases in multidrug-resistant Klebsiella pneumoniae isolates in Jordanian hospitals. J. Epidemiol. Global Health 13(2), 180–190; doi:10.1007/s44197-023-00096-2 Arafa, A.A., Hedia, R.H., Dorgham, S.M., Ibrahim, E.S., Bakry, M.A., Abdalhamed, A.M. and Abuelnaga, A.S.M. 2022. Determination of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from horses with respiratory manifestation. Vet. World 15(4), 827–833; doi:10.14202/vetworld.2022.827-833 Bedair, N.M., Sakr, M.A., Mourad, A., Eissa, N., Mostafa, A. and Khamiss, O. 2024. Molecular characterization of the whole genome of the H9N2 avian influenza virus isolated from poultry farms in Egypt. Arch. Virol. 169(5), 1; doi:10.1007/s00705-024-06018-2 Bedawy, Y.M., Homouda, S.N., Ahmed, H.A. and Abd-el Tawab, A.A. 2024. Genotyping and antibiotic resistance profile of Klebsiella pneumoniae and Corynebacterium bovis isolates recovered from clinical and subclinical mastitis milk samples. J. Adv. Vet. Res. 14(3), 349–355. Bekele, D., Dessalegn, B., Tadesse, B. and Abey, S.L. 2024. Streptococcus equi subspecies equi from strangles suspected equines: molecular detection, antibiogram profiles and risk factors. BMC Vet. Res. 20(1), 377–386; doi:10.21203/rs.3.rs-3263231/v1 Borum, A.E. 2022. Agents isolated from horses with respiratory system infection signs. Etlik Vet. Mikrobiyol. Dergisi. 33(1), 56–62; doi:10.35864/evmd.1087584 Brat, K., Sedlacek, I., Sevcikova, A., Merta, Z., Laska, K. and Sevcik, P. 2016. Imported anthropogenic bacteria may survive the Antarctic winter and introduce new genes into local bacterial communities. Polish Polar Res. 37(1), 89–104. Byomi, A., Zidan, S., Elkamshishi, M., Sakr, M., Elsify, A., Eissa, N. and Hadad, G. 2019a. Some associated risk factors with Coxiella burnetii in sheep, humans and ticks in Menoufiya governorate, Egypt. Biosci. Res. 16(S1-2), 121–138. Byomi, A., Zidan, S., Sakr, M.A., Elsify, A., Hussien, H., Eissa, N. and Hadad, G. 2019b. Coxiella burnetii infection in milk of cattle and the risk of human infection in Menoufia Governorate. JCVR 1, 140–148; doi:10.21608/jcvr.2019.65030 Byomi, A., Zidan, S., Salama, A., Elsify, A., Hadad, G. and Eissa, N. 2018. Public health implications of toxoplasmosis in animals and women in Menoufia governorate, Egypt. Assiut Vet. Med. J. 64, 120–130; doi:10.21608/avmj.2018.168920 CDC, 2019. Threats of antibiotic resistance in the United States. Available via https://www.cdc.gov/drugresistance/biggest-threats.html Chakraborty, S., Mohsina, K., Sarker, P.K., Alam, M.Z., Karim, M.I.A. and Sayem, S.A. 2016. Prevalence, antibiotic susceptibility profiles and ESBL production in Klebsiella pneumoniae and Klebsiella oxytoca among hospitalized patients. Period. Biol. 118(1), 53–58; doi:10.18054/pb.v118i1.3160 Chang, K., Lee, N.Y., Ko, W.C., Tsai, J.J., Lin, W.R., Chen, T.C., Lu, P.L. and Chen, Y.H. 2017. Identification of factors for physicians to facilitate early differential diagnosis of scrub typhus, murine typhus, and Q fever from dengue fever in Taiwan. J. Microbiol. Immunol. Infect. 50, 104–111; doi:10.1016/j.jmii.2018.01.002 Chiu, C.H., Yeh, K.M., Siu, L.K., Fung, C.P., Lin, J.C. and Chang, F.Y. 2011. Impact of age on neutrophil phagocytic reaction with different capsular serotypes of Klebsiella pneumoniae. J. Microbiol. Immunol. Infect. 44(5), 333–337; doi:10.1016/j.jmii.2010.08.013 CLSI, 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. In the CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. Colom, K., Pérez, J., Alonso, R., Fernández-Aranguiz, A., Lariño, E. and Cisterna, R. 2003. Simple and reliable multiplex PCR assay for detection of bla TEM, bla SHV and bla OXA–1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 223(2), 147–151; doi:10.1016/S0378-1097(03)00306-9 Dan, B., Dai, H., Zhou, D., Tong, H. and Zhu, M. 2023. Relationship between drug resistance characteristics and biofilm formation in Klebsiella Pneumoniae strains. Infect. Drug Resist. 16, 985–998; doi:10.2147/IDR.S396609 Deepan, G., Bhanu Rekha, V., Ajay Kumar, V.J., Kavita Vasudevan, P. and Quintoil, N. 2023. Hygienic practices and prevalence of Klebsiella pneumoniae among butchers in a beef slaughterhouse: a comprehensive study. Age 18, 05. Dunn, S.J., Connor, C. and Mcnally, A. 2019. The evolution and transmission of multi-drug resistant Escherichia coli and Klebsiella pneumoniae: the complexity of clones and plasmids. Curr. Opinion Microbiol. 51, 51–56. Eissa, N. 2024a. Fungal diseases in dogs and cats. Cambridge, MA: Academic Press, pp: 523–32. Eissa, N. 2024b. Mycoplasma, rickettsia, and chlamydia of dogs and cats. In Introduction to diseases, diagnosis, and management of dogs and cats. Ed., Rana, T. Cambridge, MA: Academic Press, pp: 489–499. Eissa, N. 2024c. Q fever (Queensland fever) In Handbook of zoonotic diseases of goats. Ed., Rana, T. Wallingford, UK: CABI. pp: 260–267; doi:10.1079/9781800622852.0022 Eissa, N. and Harb, O.H. 2023. Diseases of the ear. In Key questions in clinical farm animal medicine. Ed., Rana, T. Wallingford, UK: CABI, 2, pp: 232–276; doi:10.1079/9781800624818.0007 Eissa, N. and Nasr, M.A. 2025. Basics of water supply, quality, and purification. In Epidemiology and environmental hygiene in veterinary public health. Ed., Rana, T. Hoboken, NJ: Wiley, pp: 439–449; doi:10.1002/9781394208180.ch38 El-Khadry, S., Elbahy, N., Elkhatam, A., Eissa, N. and Zidan, S. 2025. An overview of the potential role of water as a vehicle of zoonotic protozoa. J. Curr. Vet. Res. 7(1), 234–256; doi:10.21608/jcvr.2025.421858 Elrefaey, A. and Eissa, N. 2025. Milk: a global nutrition appeal. In Handbook of milk production, quality and nutrition. Ed., Rana, T. Cambridge, MA: Academic Press, pp: 229–242; doi:10.1016/B978-0-443-24820-7.00048-1 Elsobky, Y., Eltholth, M., Abdalla, E., Eissa, N., Hadad, G., Nayel, M., Salama, A., Mousa, W., Kamal, A., Abu-Seida, A.M. and Gaidan, O.K. 2024. Spatio-temporal dynamics and risk cluster analysis of highly pathogenic avian influenza (H5N1) in poultry: advancing outbreak management through customized regional strategies in Egypt. Open Vet. J.14, 2911; doi:10.5455/OVJ.2024.v14.ill.20 Elsobky, Y., Elsify, A.M., Rabah, I.M., Mousa, W.S., Elgendy, A.A., Sabbah, K., Zaghawa, A.A., Nayel, M.A., Salam, A.A., Abu-Seida, A.M., Abdulkarim, A. and Elkamshishi, M.M. 2025. Isolation and molecular identification of MDR Escherichia coli strains isolated from mastitic cows in Egypt. Open Vet. J. 15(5), 2094–2102; doi:10.5455/OVJ.2025.v15.i5.27 El-Sokkary, R., Uysal, S., Erdem, H., Kullar, R., Pekok, A.U., Amer, F., Grgić, S., Carevic, B., El-Kholy, A., Liskova, A. and Özdemir, M. 2021. Profiles of multidrug-resistant organisms among patients with bacteremia in intensive care units: an international ID-IRI survey. Europ. J. Clin. Microbiol. Infect. Dis. 40(11), 2323–2334. Enferad, E. and Mahdavi, S. 2020. Antibiotic resistance pattern and frequency of some beta lactamase genes in Klebsiella pneumoniae isolated from raw milk samples in Iran. J. Hellenic Vet. Med. Soc. 71(4), 2455–2462; doi:10.1016/j.jhmsoc.2018.09.010 Gomaa, N. 2021. Prevalence, antimicrobial resistance, and biofilm formation of Klebsiella pneumoniae isolated from human and cows. Zagazig Vet. J. 49(1), 27–41; doi:10.21608/zvjz.2021.51857.1126 Holt, K.E., Wertheim, H., Zadoks, R.N., Baker, S., Whitehouse, C.A., Dance, D., Jenney, A., Connor, T.R., Hsu, L.Y., Severin, J. and Brisse, S. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U. S. A. 112(27), E3574–E3581; doi:10.1073/pnas.1501049112 Hu, Y., Anes, J., Devineau, S. and Fanning, S. 2021. Klebsiella pneumoniae: prevalence, reservoirs, antimicrobial resistance, pathogenicity, and infection: a hitherto unrecognized zoonotic bacterium. Foodborne Pathogens Dis. 18, 63–84; doi:10.1089/fpd.2020.2847 Huang, S., Ji, S., Wang, F., Huang, J., Alugongo, G.M. and Li, S. 2020. Dynamic changes of the fecal bacterial community in dairy cows during early lactation. AMB. Express 10, 1–9; doi:10.1186/s13568-020-01106-3 Kaczorek-Łukowska, E., Małaczewska, J., Wójcik, R., Naumowicz, K., Blank, A. and Siwicki, A.K. 2021. Streptococci as the new dominant etiological factors of mastitis in dairy cows in north-eastern Poland: analysis of the results obtained in 2013–2019. Irish Vet. J. 74, 1–6; doi:10.1186/s13620-020-00181-z Klemmer, J., Njeru, J., Emam, A., El-Sayed, A., Moawad, A.A., Henning, K., Elbeskawy, M.A., Sauter-Louis, C., Straubinger, R.K., Neubauer, H. and El-Diasty, M.M. 2018. Q fever in Egypt: epidemiological survey of Coxiella burnetii specific antibodies in cattle, buffaloes, sheep, goats and camels. PLoS One 13(2), e0192188; doi:10.1371/journal.pone.0192188 Koovapra, S., Bandyopadhyay, S., Das, G., Bhattacharyya, D., Banerjee, J., Mahanti, A., Samanta, I., Nanda, P.K., Kumar, A., Mukherjee, R., Dimri, U. and Singh, R.K. 2016. Molecular signature of extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infect. Genet. Evol. 44, 395–402; doi:10.1016/j.meegid.2016.07.032 Kumar, D. and Kumari, S.K. 2013. Klebsiella: in drinking water. Int. J. Pharmaceut. Sci. Inven. 2(12), 38–42. Léon, A., Castagnet, S., Maillard, K., Paillot, R. and Giard, J.C. 2020. Evolution of in vitro antimicrobial susceptibility of equine clinical isolates in France between 2016 and 2019. Animals 10(5), 812; doi:10.3390/ani10050812 Liu, J., Zhang, Y., Cai, J., Shao, L., Jiang, X., Yin, X., Zhao, X. and Wang, S. 2023. Clinical and microbiological characteristics of Klebsiella pneumoniae co-infections in pulmonary tuberculosis: a retrospective study. InfectDrug. Resist. 16, 7175–7185; doi:10.1016/j.indrr.2018.07.015 Loncaric, I., Cabal Rosel, A., Szostak, M.P., Licka, T., Allerberger, F., Ruppitsch, W. and Spergser, J. 2020. Broad-spectrum cephalosporin-resistant Klebsiella spp. isolated from diseased horses in Austria. Animals 10(2), 332; doi:10.3390/ani10020332 Lucchese, L., Capello, K., Barberio, A., Zuliani, F., Stegeman, A., Ceglie, L., Guerrini, E., Marangon, S. and Natale, A. 2015. IFAT and ELISA phase I/phase II as tools for the identification of Q fever chronic milk shedders in cattle. Vet. Microbiol. 179(1-2), 102–108; doi:10.1016/j.vetmic.2015.02.010 Magill, S.S., O’Leary, E., Janelle, S.J., Thompson, D.L., Dumyati, G., Nadle, J., Wilson, L.E., Kainer, M.A., Lynfield, R., Greissman, S. and Ray, S.M. 2018. Changes in prevalence of health careu–associated infections in US hospitals. New Eng. J. Med. 379(18), 1732–1744; doi:10.1056/NEJMoa1801550 Meatherall, B.L., Gregson, D., Ross, T., Pitout, J.D. and Laupland, K.B. 2009. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am. J. Med. 122(9), 866–873; doi:10.1016/j.amjmed.2009.03.034 Mohammed, A.N., Abdel-Latef, G.K., Abdel-Azeem, N.M. and El-Dakhly, K.M. 2016. Ecological study on antimicrobial-resistant zoonotic bacteria transmitted by flies in cattle farms. Parasitol. Res. 115, 3889–3896; doi:10.1007/s00436-016-5154-7 Munoz-Price, L.S. and Quinn, J.P. 2009. The spread of Klebsiella pneumoniae carbapenemases: a tale of strains, plasmids, and transposons. Clin. Infect. Dis. 49(11), 1739–1741; doi: 10.1086/648078 Myatt, R. 2017. Sputum collection and analysis: what the nurse needs to know. Nurs Stand 31(27), 40–43; doi:10.7748/ns.2017.e10228 Namratha, K.G., Sreeshma, P., Subbannayya, K., Dinesh, P.V. and Champa, H. 2015. Characterization and antibiogram of Klebsiella spp. isolated from clinical specimen in a rural teaching hospital. Sch. J. App. Med. Sci. 3(2E), 878–883. Nasr, M.A. and Eissa, N. 2025a. Maintenance of industrial hygiene. In Epidemiology and environmental hygiene in veterinary public health. Ed., Rana, T. Hoboken, NJ: Wiley, pp: 593–597;doi: 10.1002/9781394208180.ch51 Nasr, M.A. and Eissa, N. 2025b. Rural and urban pollution. In Epidemiology and environmental hygiene in veterinary public health. Ed., Rana, T. Hoboken, NJ: Wiley, pp: 405–414; doi:10.1002/9781394208180.ch35 Osman, E.A., El-Amin, N.E., Al-Hassan, L.L. and Mukhtar, M. 2021. Multiclonal spread of Klebsiella pneumoniae across hospitals in Khartoum, Sudan. J. Global Antimicrob. Resist. 24, 241–245; doi:10.1016/j.jgar.2020.12.004 Paniagua-Contreras, G.L., Bautista-Cerón, A., Morales-Espinosa, R., Delgado, G., Vaca-Paniagua, F., Díaz-Velásquez, C.E., De La Cruz-montoya, A.H., García-Cortés, L.R., Sánchez-Yáñez, M.P. and Monroy-Pérez, E. 2023. Extensive expression of the virulome related to antibiotic genotyping in nosocomial strains of Klebsiella pneumoniae. Int. J. Mol. Sci. 24, 14754; doi:10.3390/ijms241914754 Ramzy, G., Mousa, W., Gaidan, O.K., Zaghawa, A., Nayel, M., Elsify, A., Eissa, N., Abu-Seida, A.M. and Salama, A. 2025. Molecular characterization and risk factors associated with zoonotic cryptosporidiosis in bovine calves and humans in the Menoufia governorate, Egypt. Open Vet. J. 15(1), 1–12; doi:10.5455/OVJ.2025.v15.i1.26 Randall, L.P., Cooles, S.W., Osborn, M.K., Piddock, L.J.V. and Woodward, M.J. 2004. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 53(2), 208–216; doi:10.1093/jac/dkh070 Rao, K., Patel, A., Sun, Y., Vornhagen, J., Motyka, J., Collingwood, A., Teodorescu, A., Baang, J.H., Zhao, L., Kaye, K.S. and Bachman, M.A. 2021. Risk factors for Klebsiella infections among hospitalized patients with preexisting colonization. mSphere 6(3), e0013221; doi:10.1128/mSphere.00132-21 Ribeiro, M.G., De Morais, A.B.C., Alves, A.C., Bolaños, C.A.D., De Paula, C.L., Portilho, F.V.R., De Nardi Júnior, G., Lara, G.H.B., De Souza Araújo Martins, L., Moraes, L.S., Risseti, R.M., Guerra, S.T., Bello, T.S., Siqueira, A.K., Bertolini, A.B., Rodrigues, C.A., Paschoal, N.R., De Almeida, B.O., Listoni, F.J.P., Sánchez, L.F.G. and Paes, A.C. 2022. Klebsiella-induced infections in domestic species: a case-series study in 697 animals (1997–2019). Braz. J. Microbiol. 53(1), 455–464; doi:10.1016/j.brazjmicro.2019.01.004 Salman, M., Eldin, A., Eissa, N., Maher, A., Aish, A. and Moez, S. 2024a. Evaluation of the effectiveness of some essential oils against zoonotic methicillin-resistant Staphylococcus aureus isolated from dairy products and humans. J. Adv. Vet. Anim. Res. 11(2), 306; doi:10.5455/javar.2024.k778 Salman, M.B., Zin Eldin, A.I., Ata, N.S., Abd Elfatah, E.B. and Eissa, N. 2024b. Prevalence, antimicrobial resistance and risk factors of zoonotic foodborne pathogens isolated from camel meat. JCVR 6(1), 272–284; doi:10.21608/jcvr.2024.352718 Schukken, Y., Chuff, M., Moroni, P., Gurjar, A., Santisteban, C., Welcome, F. and Zadoks, R. 2012. The “other” gram-negative bacteria in mastitis: Klebsiella, Serratia, and more. Vet. Clin. Food Anim. Pract. 28(2), 239–256; doi:10.1016/j.cvfa.2012.04.001 Schwab, F., Gastmeier, P. and Meyer, E. 2014. The warmer the weather, the more gram-negative bacteria - impact of temperature on clinical isolates in intensive care units. PLoS One 9(3), e91105; doi:10.1371/journal.pone.0091105 Selmi, R., Tayh, G., Srairi, S., Mamlouk, A., Ben Chehida, F., Lahmar, S., Bouslama, M., Daaloul-Jedidi, M. and Messadi, L. 2022. Prevalence, risk factors and emergence of extended-spectrum β-lactamase producing-, carbapenem- and colistin-resistant Enterobacterales isolated from wild boar (Sus scrofa) in Tunisia. Microbial Pathog. 163, 105385; doi:10.1016/j.micpath.2021.105385 Seyedjavadi, S.S., Goudarzi, M. and Sabzehali, F. 2016. Relationship between blaTEM, blaSHV and blaCTX-M genes and acute urinary tract infections. J. Acute Dis. 5, 71–76; doi:10.1016/j.joad.2015.07.007 Shehata, A.A., Abd-Elfatah, E.B., Elsheik, H.E., Salman, M.B., Khater, A.S.I. and El-Emam, M.M.A. 2024. Epidemiological features, biochemical indices, antibiogram susceptibility profile and biofilm factor genes of Klebsiella pneumoniae isolated from bovine clinical mastitis cases. Pak. Vet. J. 44(1), 141–147; doi: 10.29261/pakvetj/2024.130 Shilpa, K., Thomas, R. and Ramyashree, A. 2016. Isolation and antimicrobial sensitivity pattern of Klebsiella pneumoniae from sputum samples in a tertiary care hospital. Int. J. Biomed. Adv. Res. 7(2), 53–57; doi:10.1016/j.ijba.2019.01.003 Shnaiderman-Torban, A., Navon-Venezia, S., Dor, Z., Paitan, Y., Arielly, H., Abu Ahmad, W., Kelmer, G., Fulde, M. and Steinman, A. 2020. Extended-spectrum β-lactamase-producing Enterobacteriaceae shedding in farm horses versus hospitalized horses: prevalence and risk factors. Animals 10(2), 282; doi:10.3390/ani10020282 Singh, A.R. 2010. Modern medicine: toward prevention, cure, well-being and longevity. Mens Sana Monogr. 8(1), 17; doi:10.4103/0973-1229.58817 Sobhy, K.E., Abd El-hafeez, A.M., Shoukry, F.A. and Refaai, E.S. 2015. Pattern of sputum bacteriology in acute exacerbations of chronic obstructive pulmonary disease. Egypt. J. Bronchol. 9, 170–177; doi:10.4103/1687-8426.158065 Sobur, A., Haque, Z.F., Sabuj, A.A., Ievy, S., Rahman, A.T., El Zowalaty, M.E. and Rahman, T. 2019. Molecular detection of multidrug and colistin-resistant Escherichia coli isolated from house flies in various environmental settings. Future Microbiol. 14, 847–858; doi:10.2217/fmb-2019-0053 Sugiyama, M., Watanabe, M., Sonobe, T., Kibe, R., Koyama, S. and Kataoka, Y. 2022. Efficacy of antimicrobial therapy for bovine acute Klebsiella pneumoniae mastitis. J. Vet. Med. Sci. 84(7), 1023–1028; doi:10.1292/jvms.21-0617 Tanwar, J., Das, S., Fatima, Z. and Hameed, S. 2014. Multidrug resistance: an emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014, 541340; doi:10.1155/2014/541340 Toledano-Tableros, J.E., Gayosso-Vázquez, C., Jarillo-Quijada, M.D., Fernández-Vázquez, J.L., Morfin-Otero, R., Rodríguez-Noriega, E., Giono-Cerezo, S., Gutkind, G., Di Conza, J., Santos-Preciado, J.I. and Alcántar-Curiel, M.D. 2021. Dissemination of bla NDM–1 gene among several Klebsiella pneumoniae sequence types in Mexico associated with horizontal transfer mediated by IncF-like plasmids. Front. Microbiol. 12, 611274; doi:10.3389/fmicb.2021.611274 Turton, J.F., Perry, C., Elgohari, S. and Hampton, C.V. 2010. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 59(5), 541–547; doi:10.1099/jmm.0.015198-0 Wheat, P.F. 2001. History and development of antimicrobial susceptibility testing methodology. J. Antimicrob. Chemother. 48, 1–4; doi:10.1093/jac/48.suppl_1.1 WHO, 2017. Global antimicrobial resistance surveillance system (GLASS) report. Geneva, Switzerland: World Health Organization. Available via https://apps.who.int/iris/bitstream/handle/10665/279656/9789241515061-eng.pdf?ua=1 Zain Eldeen, A., Younes, A.M., Bazid, A., Eissa, N.M., Salman, M. and AboElkhair, M. 2024. Influence of some herbals on cellular immunity immunostimulation in experimentally Ndv-vaccinated chicks. Egypt. J. Vet. Sci. 55(5), 1165–1174; doi:10.21608/ejvs.2024.256822.1734 Zin Eldin, A.I., Hozyen, H.F., Shafik, E. and Eissa, N. 2025. A new Egyptian approach to the antibacterial effect of zinc oxide nanoparticles on cattle zoonotic bacteria with different transmission patterns. Open Vet. J. 15(6), 2518–2531; doi:10.5455/OVJ.2025.v15.i6.24 Zin Eldin, A.I., Salman, M.B., Salah Eldin, D.A. and Eissa, N. 2023. An overview on: bacterial fish zoonas. JCVR 5(2), 196–215; doi:10.21608/jcvr.2023.320449 | ||
| How to Cite this Article |
| Pubmed Style Eissa N, Salman MB, Younes AM, Mohamed ESA, Abu-seida AM, Abdulkarim A, Eldin AIZ. One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans. Open Vet. J.. 2025; 15(9): 4219-4234. doi:10.5455/OVJ.2025.v15.i9.28 Web Style Eissa N, Salman MB, Younes AM, Mohamed ESA, Abu-seida AM, Abdulkarim A, Eldin AIZ. One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans. https://www.openveterinaryjournal.com/?mno=263452 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i9.28 AMA (American Medical Association) Style Eissa N, Salman MB, Younes AM, Mohamed ESA, Abu-seida AM, Abdulkarim A, Eldin AIZ. One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans. Open Vet. J.. 2025; 15(9): 4219-4234. doi:10.5455/OVJ.2025.v15.i9.28 Vancouver/ICMJE Style Eissa N, Salman MB, Younes AM, Mohamed ESA, Abu-seida AM, Abdulkarim A, Eldin AIZ. One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans. Open Vet. J.. (2025), [cited January 25, 2026]; 15(9): 4219-4234. doi:10.5455/OVJ.2025.v15.i9.28 Harvard Style Eissa, N., Salman, . M. B., Younes, . A. M., Mohamed, . E. S. A., Abu-seida, . A. M., Abdulkarim, . A. & Eldin, . A. I. Z. (2025) One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans. Open Vet. J., 15 (9), 4219-4234. doi:10.5455/OVJ.2025.v15.i9.28 Turabian Style Eissa, Nourhan, Marwa B. Salman, Abdelgayed M. Younes, El Shymaa A. Mohamed, Ashraf M. Abu-seida, Abdulrahman Abdulkarim, and Asmaa I. Zin Eldin. 2025. One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans. Open Veterinary Journal, 15 (9), 4219-4234. doi:10.5455/OVJ.2025.v15.i9.28 Chicago Style Eissa, Nourhan, Marwa B. Salman, Abdelgayed M. Younes, El Shymaa A. Mohamed, Ashraf M. Abu-seida, Abdulrahman Abdulkarim, and Asmaa I. Zin Eldin. "One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans." Open Veterinary Journal 15 (2025), 4219-4234. doi:10.5455/OVJ.2025.v15.i9.28 MLA (The Modern Language Association) Style Eissa, Nourhan, Marwa B. Salman, Abdelgayed M. Younes, El Shymaa A. Mohamed, Ashraf M. Abu-seida, Abdulrahman Abdulkarim, and Asmaa I. Zin Eldin. "One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans." Open Veterinary Journal 15.9 (2025), 4219-4234. Print. doi:10.5455/OVJ.2025.v15.i9.28 APA (American Psychological Association) Style Eissa, N., Salman, . M. B., Younes, . A. M., Mohamed, . E. S. A., Abu-seida, . A. M., Abdulkarim, . A. & Eldin, . A. I. Z. (2025) One health approach on zoonotic multidrug-resistant Klebsiella pneumoniae isolated from Egyptian cattle, horses, and humans. Open Veterinary Journal, 15 (9), 4219-4234. doi:10.5455/OVJ.2025.v15.i9.28 |