| Research Article | ||
Open Vet. J.. 2025; 15(9): 4735-4743 Open Veterinary Journal, (2025), Vol. 15(9): 4735-4743 Research Article Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCRMohamed Abdusalam1*, Mohamed Elbasir1, Mohamed Ashteba1, Almabrok Saeed1, Fawzi Ebrahim2, Ammar Aslougi1, Inas Alhudiri1, Salah Edin El Meshri3, Monier Sharif4 and Adam Elzagheid11Department of Genetic Engineering, Libyan Biotechnology Research Center, Tripoli, Libya 2Department of Cell Biology and Cell Culture, Libyan Biotechnology Research Center, Tripoli, Libya 3Department of Microbiology, Libyan Biotechnology Research Center, Tripoli, Libya 4Department of Biomedicine, School of Basic Sciences, Libyan Academy for Graduate Studies, Al Bayda, Libya *Corresponding Author: Mohamed Abdusalam. Department of Genetic Engineering, Libyan Biotechnology Research Center, Tripoli, Libya. Email: mabdusalam1973 [at] gmail.com Submitted: 08/05/2025 Revised: 30/07/2025 Accepted: 16/08/2025 Published: 30/09/2025 © 2025 Open Veterinary Journal
ABSTRACTBackground: Avian influenza viruses (AIVs) pose a global threat to avian and human health, with wild migratory birds being recognized as major natural reservoirs and vectors of viral dissemination. Libya, with its diverse wetlands and geographic positioning along key migratory routes, serves as a critical stopover for numerous bird species, presenting a potential hotspot for AIV transmission. Aim: This study aimed to detect the presence of type A influenza virus in wild migratory birds in eastern Libya using reverse transcription real-time polymerase chain reaction (RT-qPCR) and evaluate the role of AIV in the ecology and surveillance of the region. Methods: From June to October 2024, 72 nasopharyngeal, cloacal, and fresh fecal swabs were collected from various species of wild migratory birds across key wetlands and lakes in eastern Libya. The morphological identification and characterization of the bird were performed. Detection of the matrix gene of AIV was conducted using the SVIP-MPv2 assay for the detection of type A influenza virus with primers and probe for H5 subtype detection adapted from validated protocols. Results: Among the 72 tested samples, one cloacal swab collected from a Eurasian teal (Anas crecca) tested positive for type A influenza virus, whereas the remaining 71 samples were negative. The positive case highlights the silent circulation of AIV among asymptomatic wild birds in the region. Clinical examinations during sampling confirmed no visible signs of illness, such as respiratory distress, lethargy, or neurological symptoms in the positive teal, consistent with typical LPAI reservoir behavior in waterfowl. However, follow-up RT-qPCR testing for the H5 subtype was negative, indicating a non-H5 influenza A strain. Conclusion: The detection of AIV in a migratory bird in Libya highlights the importance of continued surveillance in wild avian populations, especially in ecologically sensitive areas along migratory pathways. Early detection through molecular diagnostics is essential for informing public health strategies and mitigating the transmission risk to domestic poultry populations. Future efforts should prioritize the continued monitoring of AIV, with particular emphasis on the detection of H7 and H9 subtypes, using a larger sample size to enhance surveillance efforts. Keywords: Avian influenza, Libya, RT-qPCR, Type A influenza virus, Wild migratory birds. IntroductionAvian influenza viruses (AIVs) are a significant global concern, affecting both domestic and wild bird populations. These viruses encompass all 16 hemagglutinin (H) and 9 neuraminidase (N) subtypes, creating diverse combinations with varying host tropisms and pathogenicity (Alexander, 2000; Olsen et al., 2006). Wild aquatic birds maintain this global genetic pool, facilitating continuous evolution and intercontinental spread through migration, highlighting their genetic diversity and global distribution. Based on the severity of this disease in avian species, AIVs are subdivided into low pathogenicity (LPAIV) and highly pathogenic (HPAIV) variants. LPAI viruses are the most common, often causing asymptomatic infections in wild birds while maintaining a vast reservoir across more than 105 species from 26 families (Slemons et al., 1974; Munster et al., 2007). In 2005, the H5N1 outbreak among wild birds in Lake Qinghai, China, underscored how migratory birds contribute to the worldwide spread of HPAIV (Chen et al., 2005; Weber et al., 2007). Avian influenza A virus primarily circulates in wild migratory waterbirds, including the coot (Fulica atra), Eurasian teal (Anas crecca), domestic goose (Anser domesticus), domestic duck (Anas platyrhynchos domesticus), and Eurasian woodcock (Scolopax rusticola). These species and varieties are considered the main natural reservoir for avian influenza A virus (Modiri et al., 2021). These species not only exhibit higher virus isolation rates, as in the case of ducks (Shortridge, 1992; Wille et al., 2023), but also tend to remain asymptomatic despite infection. (Evseev and Magor, 2019). Consequently, waterfowl facilitate persistent environmental circulation of the virus, especially when their habitats intersect agricultural lands and live bird markets. Such interactions enable these birds to act as a bridge between wild migratory populations and domestic poultry, thereby amplifying the risk of cross-species transmission (Keawcharoen et al., 2008; Meseko et al., 2010; Youk et al., 2024). The migration of wild birds from Europe to Africa is a remarkable journey, with many species passing through eastern Libya (Defos et al., 2001; Etayeb and Essghaier, 2007). This region is rich in natural resources, boasting numerous lakes, wetlands, and freshwater marshes that provide vital stopping points for migratory birds. These habitats offer abundant food and resting places, making eastern Libya a crucial area for avian survival during migration. Researchers can closely monitor the migratory routes of these birds, tracking their movements and behaviors in real-time, using global positioning system technology (Gallego-Zamorano et al., 2024; Adojaan et al., 2025). Wetlands in eastern Libya are vital stopovers for migratory birds between Europe and Africa, as these paths facilitate the spread of viruses, as asymptomatically infected birds shed viruses into shared habitats. This ecological connectivity amplifies regional transmission risks, particularly where wetlands intersect poultry farms or live markets. These data are essential for understanding the ecological significance of these wetlands and for developing effective conservation strategies to protect both the birds and their habitats. In Libya, studies and research on AIV are limited, and there is a lack of advanced studies and research pertaining to the AIV (Kammon et al., 2022). The rapid intensification of the poultry industry over recent decades has been linked to an increased incidence of respiratory disorders among commercial chickens (Agha et al., 2023). Field studies have consistently implicated the LPAI H9N2 subtype as a primary contributor to these illnesses; co-infections with agents such as Newcastle disease virus have further elevated mortality rates in affected flocks (Al-Garib et al., 2007; Fares et al., 2010). The first occurrence of LPAIV subtype H9N2 infection in commercial poultry in Libya was documented in 2006; however, the virus could not be isolated during that time. It was isolated and characterized in 2013 (Kammon et al., 2015). In Libya, the initial instance of HPAIV (H5N1) infection in a Tubrok backyard chicken purchased from a live bird market was identified in March 2014; however, the infection did not spread beyond that date, and the spread was successfully controlled (Kammon et al., 2015). For the detection of influenza viruses in public health laboratories, reverse transcription–real-time (RT-qPCR) has established itself as a vital diagnostic tool. In September 2008, the United States the Food and Drug Administration clarified the AIV RT-qPCR detection and characterization panel commonly referred to as the RT-qPCR flu panel by the Centers for Disease Control and Prevention, thereby standardizing its use for AIV diagnosis (Shu et al., 2011). In this study, we monitored multiple migratory stopover sites across eastern Libya to investigate the presence of influenza A viruses. Nasopharyngeal, cloacal, and freshly collected fecal swabs were obtained from wild migratory birds and waterfowl, and these samples were subsequently analyzed using RT-qPCR to evaluate the region’s role in AIV ecology and surveillance. Materials and MethodsSample collectionFrom June to the end of October 2024, 72 nasopharyngeal, cloacal, and fresh fecal swabs were collected from wild migratory birds and waterfowl in eastern Libya. Samples from wild birds were collected in accordance with the regular surveillance of wild birds in Libya (Berbash et al., 2012). The identification and characterization of the birds were performed based on the knowledge of local hunters who have good experience with the names and types of wild migratory birds and were also compared to the Atlas of wintering waterbirds of Libya, 2005–2010 (Berbash et al., 2012). Figure 1 shows a map showing the names of wetlands and lakes in some areas of eastern Libya from which samples were taken. Table 1 shows the types of migratory wild birds, waterfowls, and wetland areas in which these swabs were taken. Swabs were placed in viral transport media Bio-speedy vNAT®, stored in refrigerators, and immediately sent to the Libyan Biotechnology Research Center laboratory for viral RNA extraction and AIV detection. RNA extractionViral RNA was extracted from the avian cloacal swab specimens using a magnetic bead NuActor® Automatic Extractor cartridge system (Boditech Med Inc., Chuncheon-si, Gangwon-do, Republic of Korea) according to the manufacturer’s instructions. Primers and probes for the detection of avian influenzaDetection of the type A influenza virusThe primers and probe were selected following a comprehensive in-silico screening of 99,353 AIV MP sequences from the entire database of the Influenza Virus Database (Bao et al., 2008). Primer and probe sequences were optimized and selected from Nagy et al. (2021). The SVIP-MP assay version 2 contains the initial SVIP-MP-F and SVIP-MP-R primer pair (Nagy et al., 2010 integrated DNA technologies) and a novel 5’ FAM-labeled and 3′-minor groove binder (MGB)-modified probe. The primers and probes sequences used to detect type A influenza virus by real-time RT-PCR are shown in (Table 2). The primers consist of a 182-nucleotide-long amplicon. The probe is produced in the reverse complement orientation, that is, the 5’-end is cut downstream of the reverse primer. The SVIP-MPv2 assay was also optimized and validated as a one-step reaction using the QuantiTect Probe RT-PCR Kit (Qiagen). With the ultimate primer concentration (0.6 μM each) and probe (0.21 μM) in a total volume of 25 μl (20 μl reaction mix and 5 μl total nucleic acid extract).
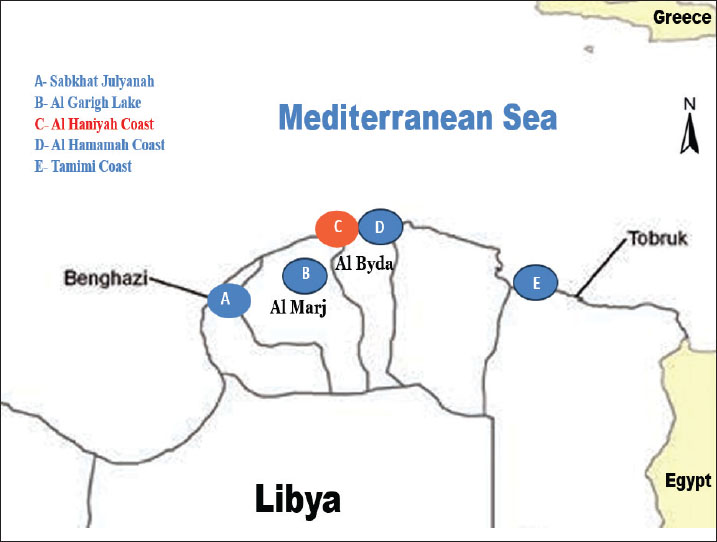
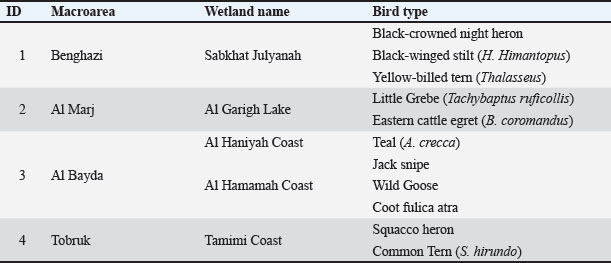
Fig. 1. A map showing the names of the wetlands and lakes in some areas of eastern Libya from which samples were taken: (A) Benghazi (Sabkhat Julyanah), (B) Al Marj (Al Garigh Lake), (C) Al Bayda (AL Haniyah Coast), (D) Al Bayda (Al Hamamah Coast), and (E) Tobruk (Al Tamimi Coast). Table 1. Types of wild migratory birds and wetland areas.
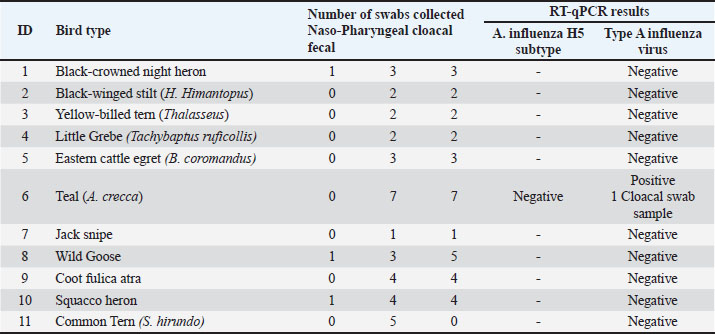
Detection of the avian influenza H5 subtypeLaboratory samples that tested positive for influenza A were retested for influenza H5 subtype by one-step real-time RT-PCR, using the primers and probe sequences that were adapted and selected from Hassan et al. (2022). The primers and probes sequences used to detect the avian influenza H5 subtype in this study are shown in (Table 3). The H5 subtype assay was developed and validated as a one-step procedure using the AgPath-ID One-Step RT-PCR Reagents Kit (Applied Biosystems). The total volume used in each reaction was 15 μl, which included 12.5 μl of master mix with primers and probes and 2.5 μl of extracted nucleic acid. Reverse transcription quantitative PCR (RT-qPCR)To identify influenza A virus in birds, we applied internationally recognized guidelines: the FAO (2023) Animal Health Standards (FAO, 2023), WOAH’s 2022 Diagnostic Manual, and Africa’s veterinary lab protocols for high-risk zoonoses. Through these protocols, a special protocol has been relied upon to diagnose the virus using real-time PCR. Firstly, to detect type A influenza virus, a PCR master mix was prepared with the QuantiTect Probe RT-PCR Kit (Qiagen). The reactions were run on a CFX 96 Touch thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA) with the following thermocycling profile: the thermal cycling protocol began with an initial reverse transcription step at 50°C for 30 minutes, followed by enzyme activation at 95°C for 15 seconds. This was succeeded by 45 amplification cycles consisting of: denaturation (95°C for 10 seconds), annealing (64°C for 30 seconds with FAM channel signal acquisition), and extension (72°C for 10 seconds). Second, for the detection of Avian Influenza H5 subtype, the PCR master mixtures were prepared with the AgPath-ID One-Step RT-PCR Reagents Kit (Applied Biosystems). The thermocycling settings were protocol started with a 10 minutes incubation at 45°C, followed by initial denaturation at 95°C for 10 seconds. Then, 45 amplification cycles were performed, each consisting of: 15-second denaturation at 95°C, 20 second annealing at 56.5°C (with FAM channel fluorescence capture), and 30-second extension at 72°C. The CFX Manager software (v3.1) automatically determined quantification cycle (Cq) values using its baseline threshold function, and the data were interpreted according to the World Health Organization (WHO) guidelines (WHO, 2016). Specimens with an appropriate RT-qPCR amplification curve and Cq value ≤38 were considered positive according to the guidelines provided by the WHO information for the molecular detection of influenza viruses (WHO, 2017). Ethical approvalThe research proposal was reviewed and approved by the Bioethics Committee at the Libyan Biotechnology Research Center (approval reference number; NBC:001. A.24.10). ResultsThe RT-qPCR test for detecting type A influenza virus was conducted on 11 wild migratory bird species. The species tested included the Black-crowned night heron, Black-winged stilt (Himantopus Himantopus), Yellow-billed tern (Thalasseus), Little Grebe (Tachybaptus), Eastern Cattle Egret (Bubulcus coromandus), Teal (A. crecca), Jacksnipe, Wild Goose, Coot, Squacco heron, and Common Tern (Sterna hirundo) (Fig. 2). Multiple swabs were taken from each bird, consisting of both nasopharyngeal and cloacal samples. Among these, only Teal tested positive for the influenza virus, with the positive result obtained from one cloacal swab. All other bird species returned negative results for Type A influenza virus at the time of sampling, indicating that they were not carrying the virus. (Table 4) showed the laboratory results for avian influenza testing in various bird species. One Teal (A. crecca) tested positive for influenza A virus but negative for the H5 subtype in the follow-up RT-qPCR test. This result points to infection with a non-H5 influenza A strain.”. Table 2. Primer and probe sequences used to detect type A influenza virus by RT-qPCR.
Table 3. The primers and probe sequences used to detect the H5 influenza subtype in this study.
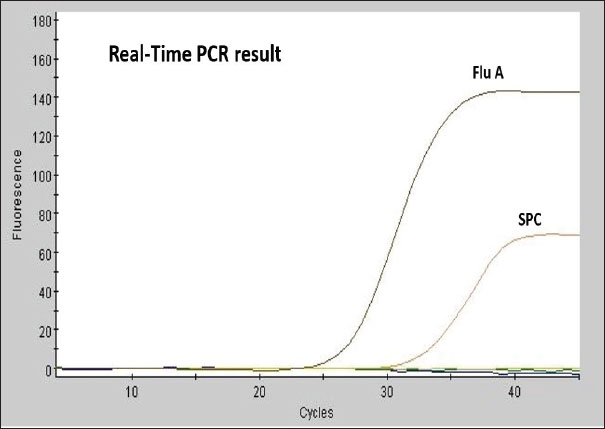
Figure 3 shows the RT-qPCR results, which measure fluorescence intensity over multiple amplification cycles. It is used to detect and quantify a target genetic sequence (likely influenza A virus in this case). Higher values indicate successful target sequence amplification. A characteristic sigmoidal amplification curve was observed, crossing the threshold at approximately Cq < 38, consistent with the presence of influenza A virus in the sample. Sample processing control curve confirms the integrity and proper performance of the PCR reaction, indicating that the reaction proceeded without inhibition and that the sample was processed correctly. DiscussionThe detection of Type A influenza virus in Teal highlights its role as a virus reservoir. Migratory birds, such as Teal, are known to carry and transmit influenza viruses over long distances, making them a key factor in the global spread of the virus (Bahl et al., 2021; Lebarbenchon et al., 2021; Wille and Barr, 2022 ). The detection of the virus in a cloacal swab indicates potential viral shedding via the gastrointestinal tract, a well-documented transmission route for avian influenza (Slusher et al., 2022). The absence of infection in the other tested species is promising, suggesting that they were not carrying the influenza virus at the time of sampling. However, continuous surveillance remains important because migratory birds can rapidly acquire and spread the virus (Pohlmann et al., 2023; Youk et al., 2024). Monitoring these birds over time is essential to detect any changes in infection status, shifts in viral prevalence, and the emergence of novel strains (WHO, 2023; Wille et al., 2023). Detection of the virus in the Teal’s cloacal swab suggests gastrointestinal shedding, a common characteristic of avian influenza infections. While nasopharyngeal swabs provide valuable data regarding respiratory infections, cloacal swabs are indispensable for identifying infections that may not show respiratory symptoms. This dual approach to sampling enhances the likelihood of detecting the virus through various shedding routes (Global Consortium for H5N1 and Related Influenza Viruses, 2022; James et al., 2023). Teals and other migratory birds serve as reservoirs for influenza viruses, affecting the geographical spread of these viruses (Ramey et al., 2023; Verhagen et al., 2024). Migratory pathway conditions and wetland habitats can facilitate virus transmission among bird populations. Understanding the ecological contexts in which these birds interact can provide insights into the dynamics of influenza transmission and guide effective management strategies (Pérez-Ramírez and Fernández, 2022). The detection of Type A influenza virus in a wild migratory bird has significant implications for both animal and public health (Häpe and Mathieu, 2022). If the virus subtype is pathogenic, such as H5N1 or H7N9, it could pose a risk to domestic poultry and, in rare cases, to humans. This underscores the importance of monitoring wild bird populations for influenza viruses and collaborating with public health and animal health authorities to mitigate potential risks (WHO, 2021; Food and Agriculture Organization of the United Nations, 2022; Swayne and Kapczynski, 2023). Further testing is needed to identify the specific subtype of the virus detected in the Teal, as this information is critical for assessing the potential risks to wildlife and livestock. Additionally, expanding surveillance to include more bird species and sampling locations would provide a better understanding of the virus’s distribution and circulation in the region. Monitoring the health of the Teal and other birds in the area is recommended to track any signs of illness or further transmission. The largely negative findings across most bird species offer a positive sign; however, the detection of AIV in Teal highlights the critical importance of continued surveillance of migratory bird populations. This finding emphasizes the need for ongoing research and cross-institutional cooperation to mitigate potential threats, protect wild and domestic avian populations, and ultimately preserve human health security. Table 4. RT-qPCR results for various bird species, indicating the number and type of swabs collected, and their respective test results.
Fig. 2. Photographs of two types of wild migratory birds. (A): Squacco heron in Tobruk (Al Tamimi Coast). (B): Black-winged stilt (H. Himantopus) in the city of Benghazi (Sabkhat Julyanah).
Fig. 3. RT-qPCR amplification plots Flu A amplification curve (positive detection) In the X-axis (Cycles): Represents the number of PCR cycles. Fluorescence signals increase exponentially when the target nucleic acid is present. In the Y-axis (Fluorescence Intensity): represents the detected fluorescence level. ConclusionThe detection of AIV from a migratory bird in Libya highlights the need for ongoing monitoring of wild birds, particularly in ecologically sensitive areas along migratory flyways. Molecular diagnostics are critical for early detection, public health action, and prevention of possible spillover to domestic poultry. The detection of a non-H5 strain underscores the silent circulation of diverse LPAI subtypes in migratory populations. Although less immediately threatening than HPAI H5, such strains contribute to viral evolution and pose risks through reassortment or adaptation to poultry. Global health agencies (WHO, OIE, and FAO) recommend integrated surveillance for all major avian flu subtypes (H5, H7, and H9) to mitigate risks to both animal and human health. Based on these recommendations, considering that the modest sample size of this study limits broader conclusions, it has become necessary to continue detecting the H7 and H9 subtypes of avian influenza in the next phases with a larger number of samples. AcknowledgmentsWe would like to express our thanks to Dr. Khaled Etayeb (Zoology Department, Faculty of Science, University of Tripoli, Libya) for his valuable assistance in providing information on the wetlands and lakes of eastern Libya, as well as the identification and classification of wild migratory bird species. We also extend our appreciation to the local hunters in eastern Libya for their assistance in the identification and characterization of wild migratory birds. We gratefully acknowledge the International Atomic Energy Agency (IAEA) for its support in providing essential diagnostic kits for virus detection. Conflict of interestThe authors declare that they have no conflict of interest. FundingThe authors declare that no funding was received for this study. Authors’ contributionsMA, IA, and AE designed the study. MA, ME, MAs, and AS conducted fieldwork in eastern Libya and collected swab samples. MA, FE, AA, SE, MS, and AE performed data analysis and contributed to drafting and revising the manuscript. All authors have read and approved the final version of the manuscript. Data availabilityAll data supporting this study’s findings are available in the manuscript, and no additional data sources are required. ReferencesAdojaan, K., Sellis, U., Väli, Ü., Ojaste, I., Denac, K., Lõhmus, A. and Ķuze, J. 2025. BirdMap Data – GPS tracking of Storks, Cranes and birds of prey, breeding in Northern and Eastern Europe. PlutoF. Occurrence dataset. Availabla via https://doi.org/10.15468/vnwmrx (Accessed 10 April 2025). Agha, A., Benlashehr, I., Naffati, K., Bshina, S. and Khashkhosha, A. 2023. Correlation of avian influenza-H9N2 with high mortality in broiler flocks in the southwest of Tripoli, Libya. Open Vet. J. 13, 715–722. Alexander, D.J. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74(00), 3–13. Al-Garib, S., Kammon, A., Asheg, A., Hamid, M., Fathalla, H. and Lawala, A. Detection of antibodies to avian influenza H9N2 in broiler flocks clinically expressing a respiratory disease syndrome. In Proceeding The 15th Congress & Exhibition of the World Veterinary Poultry Association, Beijing, China, 2007, 130, pp 1–108. Bahl, J., Guillemain, M. and Wong, P.W.S. 2021. Ecological and evolutionary dynamics of avian influenza viruses in migratory birds. Influenza Other Respir. Viruses 15(3), 299–309; doi:10.1111/irv.12814 Bao, Y., Bolotov, P., Dernovoy, D., Kiryutin, B., Zaslavsky, L., Tatusova, T. 2008. The influenzavirus resource at the National Center for Biotechnology Information. J. Virol. 82, 596–601; doi:10.1128/JVI.02005-07 Berbash, A., Hamza, A., Amadesi, B., Bouras, E., Azafzaf, H., Dlensi, H., Borg, J., Sultana, J., Yahia, J., Smart, M., Zenatello, M., Baccetti, N., Defos Du Rau, P. and Bashimam, W. 2012. Atlas of wintering waterbirds of Libya, 2005-2010. Chen, H., Smith, G.J.D., Zhang, S.Y., Qin, K., Wang, J., Li, K.S., Webster, R.G., Peiris, J.S.M. and Guan, Y. 2005. H5N1 virus outbreak in migratory waterfowl. Nature 436, 191–192. Defos, P., Essghaier M. and Etayeb, K. 2001. Preliminary survey of coastal wetlands of Libya, Office National de la Chasse et de la Faune Sauvage, France and Environment general Authority - Libya (report 1). Etayeb, K. and Essghaier, M. 2007. Breeding of marine birds on Farwa Island, western Libya. Ostrich 78, 419–421. Evseev, D. and Magor, K.E. 2019. Innate immune responses to avian influenza viruses in ducks and chickens. Vet. Sci. 6(1), 5; doi:10.3390/vetsci6010005 FAO. 2023. Veterinary laboratory testing protocols for priority zoonotic diseases in Africa. Rome: FAO Animal Production and Health Guidelines No. 34. https://doi.org/10.4060/cc3956en Fares, A.R., Elhafi, G., Alzaletny, R. and Gerish, E. 2010. Seroprevalence of avian influenza in broilers and layers, Northwestern area of Libya. In Congres Veterinaire Maghrebin, Hammamet, TN, pp 10–11. Food and Agriculture Organization of the United Nations. 2022. Global surveillance plan for influenza viruses in animals. Rome: FAO Animal Production and Health Report. Gallego-Zamorano, J., Davies, J., Reinartz, R., Robinson, R., Gargallo, G., Faverjon, C., Sierdsema, H. and Stahl, J. 2024. Updates to the wild bird abundance and movement models for the early warning system for avian influenza in the EU. EFSA Supporting Publications. 21. 10.2903/sp.efsa. 2024.EN-9000. Global Consortium for H5N1 and Related Influenza Viruses. 2022. Role of migratory birds in the intercontinental spread of avian influenza H5N1: a synthesis of evidence. Nat. Commun. 13(1), 4572. Häpe, M. and Mathieu, C. 2022. Avian influenza viruses in wild birds: a threat to animal and human health. Int. J. Infect. Dis. 124, 267–275; doi:10.1016/j.ijid.2022.10.014 Hassan, K.E., Ahrens, A.K., Ali, A., El-Kady, M.F., Hafez, H.M., Mettenleiter, T.C., Beer, M. and Harder, T. 2022. Improved subtyping of avian influenza viruses using an RT-qPCR-based low-density array: ‘Riems Influenza a Typing Array’, version 2 (RITA-2). J. Viruses 14(2), 415; doi:10.3390/v14020415 James, J., Slomka, M.J., Reid, S.M., Thomas, S.S., Mahmood, S., Byrne, A.M.P. and Brown, I.H. 2023. Comparative detection of avian influenza virus by RT-qPCR, ELISA, and lateral flow immunoassay in cloacal and oropharyngeal samples from wild birds. Transbound. Emerg. Dis. 70(1), 214–225. Kammon, A., Doghman, M. and Eldaghayes, I. 2022. Surveillance of the spread of avian influenza virus type A in live bird markets in Tripoli, Libya, and determination of the associated risk factors. Vet. World 15(7), 1684–1690. Kammon, A., Heidari, A., Dayhum, A., Eldaghayes, I., Sharif, M., Monne, I., Cattoli, G., Asheg, A., Farhat, M. and Kraim, E. 2015. Characterization of avian influenza and Newcastle disease viruses from Poultry in Libya. Avian Dis. 59(3), 422–430. Keawcharoen, J., Van Riel, D., Van Amerongen, G., Bestebroer, T., Beyer, W.E., Van Lavieren, R., Osterhaus, A.D., Fouchier, R.A. and Kuiken, T. 2008. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14(H5N1), 600–607. Lebarbenchon, C., Chaber, A.L. and Epstein, J.H. 2021. Migratory birds and the spread of influenza A viruses. Virus Evol. 7(2), 38; doi:10.1093/ve/vez038 Meseko, C.A., Oladokun, A.T., Solomon, P. and Yakubu, B. 2010. Detection of highly pathogenic avian influenza (H5N1) in apparently healthy ducks (Anas Sparsa Sparsa) in Live Bird Markets, Nigeria. Nigerian Vet. J. 31, 164–169. Modiri, H.A., Ghalyanchilangeroudi, A., Fallah M, M.H., Aghajantabar, S., Sadri, N., Ziafati, K.Z., Hojabr, R.A., Ashrafi, T.I., Haghbin, N.H., Parvandar, A.K., Tolooe, A. and Hosseini, H. 2021. Detetection of Influenza A viruses in migratory birds at live bird markets of Iran. Iranian Journal of Virology 15(2), 82–87. Munster, V.J., Baas, C., Lexmond, P., Waldenström, J., Wallensten, A., Fransson, T., Rimmelzwaan, G.F., Beyer, W.E.P., Schutten, M., Olsen, B., Osterhaus, A.D.M.E. and Fouchier, R.A.M. 2007. Spatial, temporal, and species variation in prevalence of influenza a viruses in wild migratory birds. PLoS. Pathogens 3(5), e61. Nagy, A., Černíková, L., Kunteová, K., Dirbáková, Z., Thomas, S.S., Slomka, M.J., Dán, A., Varga, T., Máté, M., Jiřincová, H. and Brown, I.H. 2021. A universal RT-qPCR assay for “One Health” detection of influenza A viruses. PLoS One 16(1), e0244669; doi: 10.1371/journal.pone.0244669 Nagy, A., Vostinakova, V., Pirchanova, Z., Cernikova, L., Dirbakova, Z., Mojzis, M. 2010. Development and evaluation of a one-step real-time RT-PCR assay for universal detection of influenza A viruses from avian and mammal species. Arch. Virol. 155, 665–673; doi:10.1007/s00705-010-0636-x Olsen, B., Munster, V.J., Wallensten, A., Waldenström, J., Osterhaus, A.D.M.E. and Fouchier, R.A.M. 2006. Global patterns of influenza A virus in wild birds. Science 312, 384–388. Pérez-Ramírez, E. and Fernández, J.G. 2022. Influenza A virus circulation in migratory waterfowl and its implications for disease ecology. Sci. Total Environ. 833, 154934; doi:10.1016/j.scitotenv.2022.154934 Pohlmann, A., King, J., Fusaro, A., Zecchin, B., Banyard, A.C., Brown, I.H. and Beer, M. 2023. Has epizootic become enzootic? Evidence for a fundamental change in the ecology of avian influenza in Europe. Nat. Commun. 14(1), 4156. Ramey, A.M., Hill, N.J., Spackman, E., Puryear, W.B., Keeler, S.P. and Prosser, D.J. 2023. Transmission dynamics of avian influenza in North American teals: experimental infection and environmental persistence. PLoS Pathog. 19(8), 1011589. Shortridge, K.F. 1992. Pandemic influenza: a zoonosis?. Semin. Respir. Infect. 7, 11–25. Shu, B., Wu, K.H., Emery, S., Villanueva, J., Johnson, R., Guthrie, E., Berman, L., Warnes, C., Barnes, N., Klimov, A. and Lindstrom, S. 2011. Design and performance of the CDC real-time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. J. Clin. Microbiol. 49(7), 2614–2619; doi:10.1128/JCM.02636-10 Slemons, R.D., Johnson, D.C., Osborn, J.S. and Hayes, F. 1974. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis. 18(1), 119–124. Slusher, M.J., Thorwa, S.A. and DeLiberto, T.J. 2022. Detection of avian influenza viruses in wild birds: a review of sampling methods. Journal of Wildlife Diseases 58(2), 259–273; doi:10.7589/JWD-D-21-00043 Swayne, D.E. and Kapczynski, D.R. 2023. Global strategies for controlling avian influenza in wild and domestic birds. Avian Diseases 67(1), 123–135; doi:10.1637/aviands-2022-00117 Verhagen, J.H., Fouchier, R.A.M., Lewis, N.S. and Kuiken, T. 2024. The role of Anas species in the global influenza A virus ecosystem. Nat. Rev. Microbiol. 22(1), 18–32. Weber, S., Harder, T., Starick, E. 2007. Molecular analysis of highly pathogenic avian influenza virus subtype H5N1 isolated from wild birds and mammals in northern Germany. J. Gen. Virol. 88, 554–558. Wille, M. and Barr, I.G. 2022. Resurgence of avian influenza virus. Science 376(6592), 459–460. Wille, M., Lisovski, S., Risely, A., Ferenczi, M., Roshier, D. and Hurt, A.C. 2023. Evolutionary dynamics and global diversity of avian influenza viruses circulating in waterfowl. PLoS. Pathogens 19(4), e1011276. World Health Organization (WHO). 2023. Avian influenza A(H5N1) – Current situation and risk assessment. Geneva: World Health Organization. Available via https://www.who.int/publications/m/item/avian-influenza-a(h5n1)-risk-assessment World Health Organization 2021. Influenza at the human-animal interface: summary and assessment, 1 October 2020 to 31 March 2021. Weekly Epidemiol. Record 96(23), 265–280. World Health Organization. 2016. Laboratory procedures for detection of influenza virus in specimens from suspected human cases. Geneva: World Health Organization. World Health Organization. 2017. WHO information for the molecular detection of influenza viruses. Geneva: World Health Organization. Youk, S., Torchetti, M.K., Lantz, K., Lenoch, J.B., Killian, M.L., Leyson, C. and Swayne, D.E. 2024. Highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus in wild birds, United States, 2021–2022. Emerg. Infect. Dis. 30(3), 547–551. | ||
| How to Cite this Article |
| Pubmed Style Abdusalam M, Elbasir M, Ashteba M, Saeed A, Ebrahim F, Aslougi A, Alhudiri I, Meshri SEE, Sharif M, Elzagheid A. Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR. Open Vet. J.. 2025; 15(9): 4735-4743. doi:10.5455/OVJ.2025.v15.i9.78 Web Style Abdusalam M, Elbasir M, Ashteba M, Saeed A, Ebrahim F, Aslougi A, Alhudiri I, Meshri SEE, Sharif M, Elzagheid A. Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR. https://www.openveterinaryjournal.com/?mno=256952 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i9.78 AMA (American Medical Association) Style Abdusalam M, Elbasir M, Ashteba M, Saeed A, Ebrahim F, Aslougi A, Alhudiri I, Meshri SEE, Sharif M, Elzagheid A. Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR. Open Vet. J.. 2025; 15(9): 4735-4743. doi:10.5455/OVJ.2025.v15.i9.78 Vancouver/ICMJE Style Abdusalam M, Elbasir M, Ashteba M, Saeed A, Ebrahim F, Aslougi A, Alhudiri I, Meshri SEE, Sharif M, Elzagheid A. Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR. Open Vet. J.. (2025), [cited January 25, 2026]; 15(9): 4735-4743. doi:10.5455/OVJ.2025.v15.i9.78 Harvard Style Abdusalam, M., Elbasir, . M., Ashteba, . M., Saeed, . A., Ebrahim, . F., Aslougi, . A., Alhudiri, . I., Meshri, . S. E. E., Sharif, . M. & Elzagheid, . A. (2025) Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR. Open Vet. J., 15 (9), 4735-4743. doi:10.5455/OVJ.2025.v15.i9.78 Turabian Style Abdusalam, Mohamed, Mohamed Elbasir, Mohamed Ashteba, Almabrok Saeed, Fawzi Ebrahim, Ammar Aslougi, Inas Alhudiri, Salah Edin El Meshri, Monier Sharif, and Adam Elzagheid. 2025. Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR. Open Veterinary Journal, 15 (9), 4735-4743. doi:10.5455/OVJ.2025.v15.i9.78 Chicago Style Abdusalam, Mohamed, Mohamed Elbasir, Mohamed Ashteba, Almabrok Saeed, Fawzi Ebrahim, Ammar Aslougi, Inas Alhudiri, Salah Edin El Meshri, Monier Sharif, and Adam Elzagheid. "Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR." Open Veterinary Journal 15 (2025), 4735-4743. doi:10.5455/OVJ.2025.v15.i9.78 MLA (The Modern Language Association) Style Abdusalam, Mohamed, Mohamed Elbasir, Mohamed Ashteba, Almabrok Saeed, Fawzi Ebrahim, Ammar Aslougi, Inas Alhudiri, Salah Edin El Meshri, Monier Sharif, and Adam Elzagheid. "Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR." Open Veterinary Journal 15.9 (2025), 4735-4743. Print. doi:10.5455/OVJ.2025.v15.i9.78 APA (American Psychological Association) Style Abdusalam, M., Elbasir, . M., Ashteba, . M., Saeed, . A., Ebrahim, . F., Aslougi, . A., Alhudiri, . I., Meshri, . S. E. E., Sharif, . M. & Elzagheid, . A. (2025) Monitoring influenza A virus in wild migratory birds and waterfowl in Libya using RT-qPCR. Open Veterinary Journal, 15 (9), 4735-4743. doi:10.5455/OVJ.2025.v15.i9.78 |