| Research Article | ||
Open Vet. J.. 2025; 15(9): 4393-4402 Open Veterinary Journal, (2025), Vol. 15(9): 4393-4402 Research Article Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, NigeriaEmmanuel Nnabuike Ugbo1,2, Mustofa Helmi Effendi2,3*, Agatha Ifunanya Ugbo4, Valentine Nnachetam Unegbu5, Bernard Nnabuife Agumah1, Hartanto Mulyo Raharjo6, Wiwiek Tyasningsih6, John Yew Huat Tang3, Budiastuti Budiastuti7 and Saifur Rehman81Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, Nigeria 2Division of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 3School of Food Industry, Faculty of Bioresources, and Food Industry, Universiti Sultan Zainal Abidin (Besut Campus), Besut, Malaysia 4Department of Microbiology and Parasitology, David Umahi Federal University of Health Sciences, Uburu, Nigeria 5Department of Biological Sciences, University of Agriculture and Environmental Sciences, Umuagwo, Nigeria 6Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 7Study Program of Pharmacy Science, Faculty of Health Science, Universitas Muhammadiyah Surabaya, Surabaya, Indonesia 8Department of Pathobiology, Faculty of Veterinary and Animal Sciences, Gomal University, Dera Ismail Khan, Pakistan *Corresponding Author: Mustofa Helmi Effendi. Division of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia. Email: mustofa-h-e [at] fkh.unair.ac.id Submitted: 21/04/2025 Revised: 22/07/2025 Accepted: 11/08/2025 Published: 30/09/2025 © 2025 Open Veterinary Journal
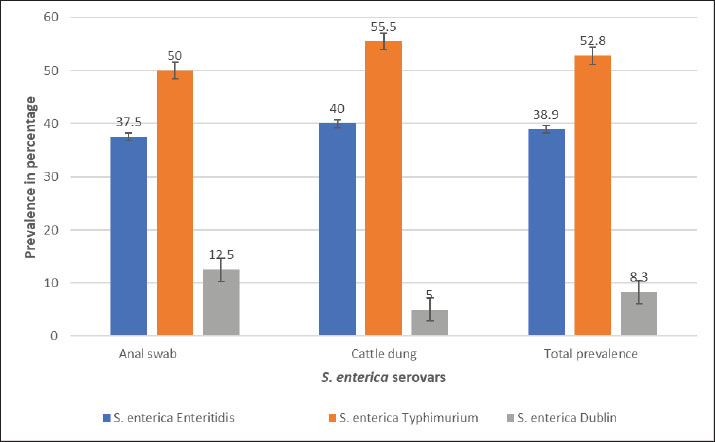
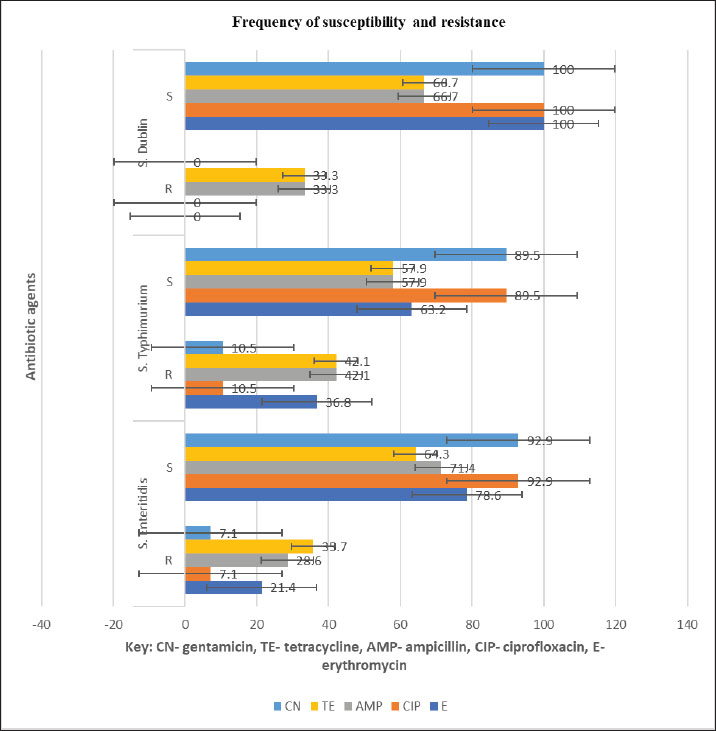
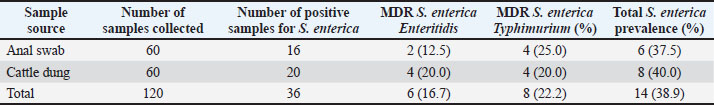
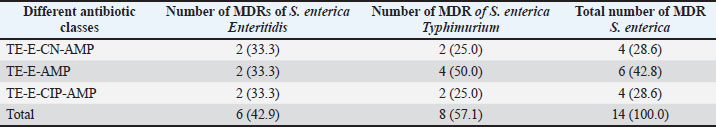
AbstractBackground: Salmonella is an important zoonotic pathogen and a causative agent of salmonellosis. Foodborne diseases have become a significant global issue that impacts food safety and public health. Aim: This study aimed to assess the public health impact of multidrug-resistant Salmonella enterica serovars identified by Matrix-Assisted Laser Desorption–Ionization Time-of-flight mass spectrometry (MALDI-TOF MS) from cattle at abattoirs. Methods: A total of 120 samples, including anal swabs and cattle dung, were collected and analyzed using standard microbiological techniques. Serovars of S. enterica were detected using MALDI-TOF MS. Antimicrobial sensitivity tests were performed on the identified isolates using the Kirby–Bauer diffusion method. Results: Of the 120 samples studied, 36 (30.0%) were confirmed to harbor S. enterica serovars. Enteritidis was identified in 14 (38.9%), Salmonella typhimurium 19 (52.8%), and S. dublin 3(8.3%). Multidrug-resistant was reported in 14 (38.9%); S. enteritidis was recorded in 6 (16.7%), and S. typhimurium had 8 (22.2%). The isolated S. enterica serovars had a resistance range of 33.3%–42.1% for ampicillin and tetracycline and a susceptibility range of 89.5%–100.0% for ciprofloxacin and gentamicin. Three distinct patterns of the multidrug resistance phenotype in S. enterica serovars have been reported. Conclusion: This study identified the presence of multidrug-resistant S. enterica serovar isolates, and if left unchecked, could lead to a public health threat. Ciprofloxacin and gentamicin were effective against the isolates and can be used to treat zoonotic foodborne salmonellosis infection. Thus, antibiotic stewardship is important to monitor resistance among bacterial organisms. Keywords: Public health, MDR, MALDI-TOF MS, S. enterica serovars, Abattoir. IntroductionSalmonella causes millions of foodborne infections and thousands of deaths each year (Galán-Relaño et al., 2023). The growing resistance of Salmonella to antibiotics in animals, food sources, and the environment, which is deemed critically important by the World Health Organization, raises serious concerns for public health and the future effectiveness of essential treatments (Wang et al., 2024). Salmonella can spread to humans through contaminated food or water, as well as through direct interaction with animals or their waste (Centers for Disease Control and Prevention (CDC), 2021). The demand for and use of animal waste products in enhancing crop growth on the farm is increasing in underdeveloped countries, and this poses a serious public health threat (Widodo et al., 2022; Khairullah et al., 2025). Livestock feces are heavily populated with Enterobacteriaceae, such as Salmonella and Escherichia coli pathogens, which are commensal flora of the gastrointestinal tract (Pradika et al., 2019; Laitinen et al., 2020; Yanestria et al., 2022). Effective host-to-host transmission enhances the spread and persistence of pathogens within host populations (Mahvish et al., 2020; Yunita et al., 2020). Pathogens bridge the immunity of a host to establish infection in a susceptible host (McLaren and Callahan, 2020). The degree of colonization and transmission is influenced by a combination of intrinsic and acquired traits from both the host and the pathogen (Oludiaro et al., 2020; Khairullah et al., 2022). Salmonella bacterium initiates infection in salmonellosis, and successful transmission is shaped by a complex interplay of pathogen virulence factors and host susceptibility mechanisms (Tanner and Kingsley, 2018). Salmonella is a zoonotic, foodborne bacterium and the causative agent of salmonellosis. In humans, the primary route of transmission is faecal-oral, though transovarial transmission has been documented in poultry (Al-Ansari et al., 2021). The most common exposure occurs through the consumption of food or water contaminated with human or animal feces, including raw fruits and vegetables (Oludiaro et al., 2022). Salmonella attachment to food is a prerequisite for colonization and subsequent transmission to humans and animals. Once attached, the bacterium is difficult to remove, even through washing (World Health Organization, 2022). Several serovars of S. enterica, such as S. Enteritidis, S. Typhimurium, and S. Senftenberg, exhibit strong adhesion to foods contaminated by animal/human feces (Abatcha et al., 2020; Elpers et al., 2020). Person-to-person transmission is also possible, especially in households and among young children in day-care settings, and oral-anal contact can contribute to transmission (New Jersey Department of Health, 2017). In poultry, vertical transmission occurs when S. Enteritidis colonizes reproductive organs, enabling the transfer of bacteria from hens to eggs or chicks (Shen et al., 2022). In addition, vectors, including arthropods and fomites, can facilitate the spread of Salmonella from contaminated environments to humans, and vectors act as vehicles for pathogen transfer, increasing the risk of exposure (Yanestria et al., 2019; Cocciolo et al., 2020; Rizzo et al., 2021). The vectors can transmit Salmonella both externally and internally, thus contributing to broader public health concerns (Oludiaro et al., 2023). Emergence of antimicrobial-resistant Salmonella strains isolated from fecal sources has become a significant public health concern. AMR in pathogens often leads to more severe infections, with increased hospitalization duration, higher treatment costs, and reliance on less effective or more toxic drugs (Awofisayo-Okuyelu et al., 2019). Infections caused by resistant Salmonella have been linked to elevated morbidity and mortality, particularly among immunocompromised individuals. The presence of resistant Salmonella in the environment is a threat to public health (Wibisono et al., 2020). Resistance mechanisms in Salmonella include enzymatic degradation of antibiotics, modification of drug target sites, reduced membrane permeability, efflux pump activity, and horizontal gene transfer. These bacteria can also disseminate resistance genes to other microbial species, thereby sustaining the spread of AMR across the food chain. In Nigeria, multidrug-resistant (MDR) Salmonella species have been identified in a variety of sources, including water, farm produce, the environment, abattoirs, and poultry settings (Babatunde et al., 2017; Ugbo et al., 2023). Salmonella is a highly diverse genus with different serotypes exhibiting varying levels of pathogenicity in humans (World Health Organization, 2018). Serotypes are more frequently associated with foodborne outbreaks. Surveillance of Salmonella prevalence in animal and human populations is critical for informing public health strategies for prevention and control (Laitinen et al., 2020; Fanissa et al., 2022). The spread of Salmonella through animal feces is influenced by factors such as cattle density, feed type, farm management practices, and environmental conditions such as temperature and humidity (World Health Organization, 2018). MALDI-TOF MS has recently been used for easy and fast identification of bacteria. Bacterial identification at the genus and species level can be achieved through biomarker detection or by matching MS profiles using the MALDI-TOF MS database (Topalcengiz et al., 2020). Therefore, this study aimed to assess the public health impact of multidrug-resistant S. enterica serovars identified via MALDI-TOF MS from cattle in Abakaliki abattoirs, Nigeria. Materials and MethodsFresh cattle dung and anal swabs of ready-to-sale healthy cattle in an abattoir within the Abakaliki urban area were the samples studied. This study used a cross-sectional design. The sample collection period was from January 2025 to March 2025. Sample collectionExactly 120 samples were collected from anal swabs and dung of cattle about to be slaughtered. Sixty (60) samples were collected from the anal and dung of cattle from two major slaughterhouses in Abakaliki. The cattle dung samples were taken aseptically using a sterile swab stick by the stabbing technique, and the sample was immersed in a test tube containing 10 ml of buffered peptone water. The anal swabs were collected by inserting 3 cm into the anus of the cattle, rotated to an angle of 360o and were deep into a test tube containing 10 ml buffered peptone water. The test tubes were labeled and stored in an icebox. Within an hour of collection, the samples were transported to the Department of Applied Microbiology Laboratory of Ebonyi State University, Abakaliki, for bacteriological analysis. The samples were examined immediately upon arrival at the laboratory and further studied for the presence of Salmonella organisms. Standard microbiological quality control measures were followed during sample collection and analysis. Preliminary isolation and identification of Salmonella speciesAnal and dung swab samples were collected and inoculated into 5 ml of selenite F broth (Oxoid, UK) for pre-enrichment, followed by overnight incubation. After incubation, a sterile wire loop was used to transfer a loopful of the culture onto SS agar (Sigma Aldrich, Germany). Plates were incubated aerobically at 37°C for 18–24 hours. Suspected Salmonella colonies appeared as non-lactose fermenters with characteristic black centers on SS agar. To obtain pure cultures, the colonies were subcultured on nutrient agar using the streak plate method. A distinct colony was picked with a sterilized wire loop and streaked across a new nutrient agar plate in a zigzag pattern, and then incubated at 37°C. The resulting pure isolates were preserved for further analysis. Presumptive Salmonella isolates were subjected to Gram staining and evaluated based on morphological and biochemical characteristics, including hydrogen sulfide (H2S) production, citrate utilization, urease activity, indole production, motility, and sugar fermentation tests for confirmation (Ugbo et al., 2023). Detection of S. enterica serovars using MALDI-TOF MS assayPure colonies of overnight-grown, unidentified Salmonella isolates were collected using Amies agar gel transport swabs with charcoal (Thermo-Fisher Scientific, Germany) and immediately sent for species identification via MALDI-TOF MS analysis. A single colony from each pure culture plate was transferred into a 1.5 mL Eppendorf tube, suspended in 300 µl of high-performance liquid chromatography (HPLC)-grade water, and thoroughly homogenized using a vortex mixer. To inactivate the bacterial cells, 900 µl of absolute ethanol was added to each tube, followed by another round of vortexing. Protein extraction was performed following previously described protocols (Persad et al., 2022; Murugaiyan et al., 2024). The inactivated bacterial suspension was centrifuged at 11,000 × g for 2 minutes, and the residual ethanol was removed from the resulting pellet. The pellets were then resuspended in 50 µl of 70% formic acid and 50 µl of acetonitrile. The suspension was sonicated at 100% amplitude with a duty cycle of 1.0 for 1 min on ice and then centrifuged at 11,290 × g for 5 minutes at room temperature. Clear supernatants were collected for MALDI-TOF MS analysis. A 1 µl aliquot of each extract was spotted onto a polished steel target plate (MSP 96 target; Bruker Daltonics, Bremen, Germany), air-dried, and subsequently overlaid with 1 µl of a saturated α-cyano-4-hydroxycinnamic acid (HCCA) matrix solution (50% acetonitrile, 0.25% trifluoroacetic acid). MALDI-TOF MS measurements were performed using the Microflex LT system (Bruker Daltonics, Bremen, Germany) and analyzed using MBT Compass Explorer version 4.1. Bacterial species were identified based on the log score values (0–3.0) according to the manufacturer’s criteria. Isolates scoring ≥2.300 were considered reliably identified, while those with scores below 2.300 were excluded from further analysis. Antimicrobial susceptibility testing of S. enterica serovar isolatesAn antimicrobial susceptibility test was performed using the Kirby–Bauer disc diffusion method to evaluate the response of S. enterica serovar isolates to the following antibiotics: ampicillin (AMP, 10 μg), gentamicin (CN, 30 μg), erythromycin (E, 15 μg), ciprofloxacin (C, 5 μg), and tetracycline (TE, 30 μg) (Oxoid, UK). MHA was prepared following the manufacturer’s instructions. A 0.5 McFarland standard suspension of each isolate was inoculated onto the agar surface using a sterile swab stick to ensure uniform lawn growth. Five antibiotic-impregnated disks were then aseptically placed on each 90-mm Petri dish, with a 30-mm spacing between disks and 15 mm from the edge of the plate. The plates were left at room temperature for 10–15 minutes to allow antibiotic diffusion, then inverted and incubated at 37°C for 18–24 hours. The zones of inhibition surrounding each disc were measured using a digital caliper following incubation, and the results were interpreted as susceptible or resistant according to the Clinical and Laboratory Standards Institute (CLSI, 2018) guidelines. Data analysesThe chi-square test was used to determine statistically significant associations between the multidrug-resistant S. enterica serovars and the sample sources. Statistical significance was set at p < 0.05. Ethical approvalThe ethical clearance was not applicable. Oral consent was obtained from the cattle owner in the abattoirs. ResultsA total of 120 samples (anal swab, 60; cattle dung, 60) were collected from an abattoir in Abakaliki and studied for the presence of S. enterica serovar isolates. Of the 120 samples, 36 (30.0%) were confirmed to harbor S. enterica serovar isolates, 16 (26.7%) were anal swab samples, and 20 (33.3%) were cattle dung samples (Table 1). The prevalence of S. enterica serovar was identified as follows: Enteritidis, 14 (38.9%): 6 (37.5%) from anal swab, 8 (40.0%) from cattle dung; S. Typhimurium 19 (52.8%): 8 (50.0%) from anal swab, 11 (55.5%) from cattle dung and S. Dublin 3 (8.3%): 2 (12.5%) from anal swab, 1 (5.0%) from cattle dung (Fig. 1). The S. enterica serovar isolates exhibited varying resistance profiles to the tested antibiotics. The isolated S. enterica serovar had a resistance range of 33.3–42.1% for ampicillin and tetracycline, a resistance range of 21.4%–36.8% for erythromycin, and a susceptibility range of 89.5%–100.0% for ciprofloxacin and gentamicin (Fig. 2). Multidrug resistance was reported in 14 patients (38.9%); S. Enteritidis was recorded in 6 (16.7%), and S Typhimurium had 8 (22.2%). Of the 16 S. enterica serovars isolated from anal swabs, 2 (12.5%) and 4 (25.0%) were MDR S. Enteritidis and S. Typhimurium, respectively. Of the 20 S. enterica serovars obtained from cattle dung, 4 (20.0%) were MDR S. enteritidis and S. typhimurium, and MDR were not reported in S. enterica Dublin (Table 2). The MDRs of Enteritidis and Typhimurium were significantly associated (p < 0.05) with sample sources. Three major patterns of multidrug resistance phenotype were observed from S. enterica serovar isolates, including TE-E-CN-AMP (four isolates; two each), TE-E-AMP (six isolates; two and four), and TE-E-CIP-AMP (four isolates; two each) for S. enterica serovar. Enteritidis and S. Typhimurium (Table 3). Table 1. Occurrence of Salmonella isolates in cattle from the Abakaliki abattoirs.
Fig. 1. Prevalence of S. enterica serovar isolates in cattle from the Abakaliki abattoirs DiscussionFoodborne diseases are a global concern, particularly in children and immunocompromised adults. Several Salmonella enterica serovars, including S. enteritidis, S. typhimurium, and S. dublin, were identified from cattle using MALDI-TOF MS. Previous studies have also reported various S. enterica serovars, such as S. Enteritidis, S. Typhimurium, S. Virchow, S. Infantis, and S. Hadar, in both humans and animals, all linked to foodborne illness outbreaks detected through MALDI-TOF MS (Khater et al., 2021; Persad et al., 2022). A One Health approach study in Lagos, Nigeria, identified different types of S. enterica serovars in humans, animals, and the environment via MALDI-TOF MS (Akinyem et al., 2023). Among the 36 confirmed S. enterica serovars Enteritidis accounted for 14 (38.9%), S. typhimurium for 19 (52.8%) patients, and S. Dublin for 3 (8.3%). Contamination of beef and its products with S. enterica serovars, particularly S. typhimurium, has been reported in previous studies (Ferrari et al., 2019). Typhimurium in chicken, beef, milk, eggs, and feed pellets was confirmed using MALDI-TOF MS in a previous study (Al-Amin et al., 2022). Other reports have indicated 6.0% isolation rates of S. typhimurium from fish, and chicken Enteritidis has also been detected in chickens, suggesting the presence of S. typhimurium is a prevalent pathogen in poultry farms and their products (El Sharkawy et al., 2017; Tarabee et al., 2017; Khater et al., 2021).
Fig. 2. Antibiotic susceptibility and resistance profiles of S. enterica serovar isolates in cattle from Abakaliki abattoirs. For many years, S. enterica serovars have been responsible for gastrointestinal infections in humans globally (Lee et al., 2015). These infections are acquired through animal contact, environmental exposure, and contaminated food or water consumption (Wibisono et al., 2021; Persad et al., 2022). Enteritidis and S. typhimurium are particularly associated with zoonotic foodborne diseases (Chlebicz and Śliżewska, 2018; Paniel and Noguer, 2019). Salmonella spp. causing foodborne salmonellosis has a serious impact on public health because it can be contracted from animal food products to people, and the disease caused by it can be severe and sometimes lead to loss of life and economic loss. This study also reported that the presence of S. dublin in cattle poses a public health risk because infection with this serotype can lead to invasive and life-threatening diseases in humans. Dairy calves were found to harbor 5.6% S. sativa. Dublin (Pharo et al., 2025). Another study confirmed that S. dublin as an emerging pathogen in cattle (Fritz et al., 2022). Identification of S. enterica serovars, such as S. enteritidis, in animals, such as cattle, is critical for controlling foodborne epidemics because these animals often do not exhibit noticeable signs of illness, making detection challenging. Table 2. Prevalence of multidrug-resistant S. enterica serovar isolates in cattle from Abakaliki abattoirs.
Table 3. Multidrug resistance phenotypic profile of S. enterica serovar isolates in cattle from Abakaliki abattoirs.
The S. enterica serovars exhibited varying antibiotic resistance profiles. S. enteritidis and S. typhimurium exhibited a resistance range of 33.3%–42.1% to ampicillin, tetracycline, and erythromycin, whereas S. dublin showed 33.3% resistance to these antibiotics, but 89.5%–100.0% were susceptible to ciprofloxacin and gentamicin. Salmonella isolates from humans, cattle, and abattoir environments have been reported to exhibit resistance to ampicillin, tetracycline, and erythromycin of up to 40.0% or above and to harbor beta-lactamase (bla), tet, and erm resistance genes (Ofori et al., 2023; Aworh et al., 2024; Rahman et al., 2024; Temesgen et al., 2025). The findings of this study align with those of previous studies, where S. dublin isolates from cattle showed tetracycline resistance and carried the tetA gene (Srednik et al., 2021). Studies on S. enteritidis and S. typhimurium from dairy cattle, raw meat, and other animals have been reported to be 75.0%–95.0% susceptible to ciprofloxacin and gentamicin (Parolini et al., 2024; Rahman et al., 2024; Cobo-Angel et al., 2025), which is consistent with the findings of this study. The observed susceptibility of S. enterica serovars to ciprofloxacin and gentamicin offers potential treatment options, provided that these antibiotics are not abused/overused, which could lead to the development of resistance in the bacterial organisms. MDR was detected in 14 (38.9%) of the S. enterica serovars, with 6 (16.7%) MDR S. enteritidis and 8 (22.2%) MDR S. typhimurium, whereas non-MDR S. Dublin. These findings agree with previous reports of MDR Serovars from S. enterica in cattle (Fritz et al., 2022). In Africa, the prevalence of MDR S. enterica serovars in cattle and the abattoir environment ranges from 27.6% to 58.6% in Nigeria (Aworh et al., 2024) and 53.7% to 66.6% in Ethiopia (Amare et al., 2024). A different study reported a MDR of 64.0% in S. enteritidis and S. typhimurium from livestock (Rahman et al., 2024) has a higher prevalence than that in this study. The increasing prevalence of MDR S. enterica serovars in both humans and animals has been widely documented (Carroll et al., 2017; Davidson et al., 2018). Dublin is frequently associated with septicemia in cattle (Salaheen et al., 2020). This study underscores the potential public health risk of animal meat in Abakaliki, which may transmit Salmonella from animal products to humans through the food chain. Thus, if not monitored frequently in food chains, MDR Salmonella strains could cause public health risks such as severe diarrhea in humans that could lead to death. Limitations of the studyAlthough this study reported the prevalence of MDR S. enterica serovars in cattle, some limitations must be recognized for future investigation. The study had a limited sample size and a short sampling period, seasonal variation was not considered, not all antibiotic classes were tested, and serological typing using the Kauffmann–White–Le Minor scheme or polymerase chain reaction (PCR)-based methods was not used to validate serotype identification. Thus, additional efficient molecular techniques, such as polymerase chain reaction (PCR) and whole-genome sequencing (WGS), are required in future studies. ConclusionMALDI-TOF MS identified three S. enterica serovars: S. enteritidis, S. typhimurium, and S. dublin from ready-to-slaughter healthy cattle at the Abakaliki abattoir. The MDR was reported in the following S. enterica serovars: Enteritidis and S. typhimurium. The presence of MDR S. enterica serovars poses a significant public health threat in humans due to their potential to cause foodborne illnesses, such as gastroenteritis. This study detected S. Dublin in cattle, which can persist in the animals without showing symptoms. However, when transmitted through the food chain, it can lead to severe diarrhea in humans, possibly resulting in death. Surveillance plans that monitor prudent antibiotic use in livestock management are required. Policymakers from both human and veterinary fields should restrict the use of antibiotics. The One Health approach is advocated to facilitate comprehensive monitoring and understanding of the global spread of AMR. AcknowledgmentsThe authors would like to thank the Universitas Airlangga and Ebonyi State University. FundingThe authors would like to thank Ebonyi State University, Abakaliki, Nigeria, and Lembaga Penelitian dan Pengabdian Masyarakat Universitas Airlangga, Indonesia, for their support. This study was partly supported by the International Research Consortium, Lembaga Penelitian dan Pengabdian Masyarakat, Universitas Airlangga, Surabaya, Indonesia. Year 2025 with grant number: 478/B/UN3.FKH/PL.14.01/2025. Author’s contributionsENU and AIU: Conceived, designed, and coordinated the study. BNA and VNU: designed data collection tools, supervised the field sample and data collection, and performed laboratory work as well as data entry. WT, HMR, JYHT, SR, and MHE: validation, supervision, and formal analysis. ENU and MHE: Contributed reagents, materials, and analysis tools. ENU, AIU, BB, and MHE: Carried out the statistical analysis and interpretation and participated in the preparation of the manuscript. All authors have read, reviewed, and approved the final version of the manuscript. Conflict of interestThe authors declare no conflict of interest. Data availabilityAll data are available in the revised manuscript. ReferencesAbatcha, M.G., Goni, M.D., Abbas, M.A., Jalo, I.M. and Mohammed, G. 2020. Listeria and Salmonella: an update on description, characteristics, incidence, and antibiotic susceptibility. Adv. Ani. Vet. Sci. 8(11), 1232–1249. Akinyemi, K.O., Fakorede, C.O., Linde, J., Methner, U., Wareth, G., Tomaso, H. and Neubauer, H. 2023. Whole genome sequencing of Salmonella enterica serovars isolated from humans, animals, and the environment in Lagos, Nigeria. BMC. Microbiol. 23(164), 164. Al-Amin, M.D., Mizanur, R., Marufa, A. and Mostofa, K. 2022. Development and verification of a MALDI-TOF MS-based method for rapid and confirmatory identification of Salmonella in feed and foods. Int. J. Microbiol. Biotechnol. 7(2), 84–92. Al-Ansari, M.M., Aljubali, M.M., Somily, A.M., Albarrag, A.M. and Masood, A. 2014. Isolation and molecular characterization of multidrug-resistant Salmonella enterica serovars. J. Infect. Pub. Health 14(12), 1767–1776. Amare, A., Asnakew, F., Asressie, Y., Guadie, E., Tirusew, A., Muluneh, S., Awoke, A., Assefa, M., Ferede, W., Getaneh, A. and Lemma, M. 2024. Prevalence of multidrug resistance Salmonella species isolated from clinical specimens at University of Gondar comprehensive specialized hospital Northwest Ethiopia: a retrospective study. PLoS. One 19(5), e0301697; doi:10.1371/journal.pone.0301697 Awofisayo-Okuyelu, A., Pratt, A., McCarthy, N. and Hall, I. 2019. Within-host mathematical modeling of the incubation period of S. typhi. R. Soc. Open Sci. 116(9), 182143–182158. Aworh, M.K., Nilsson, P., Egyir, B., Owusu, F.A. and Hendriksen, R.S. 2024. Rare serovars of non-typhoidal Salmonella enterica isolated from humans, cattle, and cattle abattoirs in Nigeria. PLoS. One 19(1), 296971. Babatunde, S.K., Kolawole, D.O., Adedayo, M.R., Ajiboye, A.E., Ajao, A.T. and Mustapha,O.N. 2004. Prevalence and characterization of Salmonella isolates from Ilorin poultry farms. Niger. J. Life Sci. 4, 1–4. Carroll, L.M., Wiedmann, M., Den Bakker, H., Siler, J., Warchocki, S., Kent, D., Lyalina, S., Davis, M., Sischo, W., Besser, T., Warnick, L.D. and Pereira, R.V. 2017. Whole-genome sequencing of drug-resistant Salmonella enterica isolates from dairy cattle and humans in New York and Washington states reveals source and geographic associations. .Appl. Environ. Microbiol. 83, 140. Centers for Disease Control and Prevention (CDC). 2021. Salmonella. Available via https://www.cdc.gov/Salmonella/index.html Chlebicz, A. and Śliżewska, K. 2018. Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: a review. Inter. Int. J. Environ. Res. Public Health 15, 863. Clinical and Laboratory Standards Institute. 2018. M100 Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Wayne City, PA: Clinical and Laboratory Standards Institute. . Cobo-Angel, C., Craig, M., Osman, M., Cummings, K.J. and Cazer, C.L. 2025. Regulations for antimicrobial use are associated with increased susceptibility among bovine Salmonella isolates from a U.S. surveillance system. One Health 20, 100983–100995. Cocciolo, G., Circella, E., Pugliese, N., Lupini, C., Mescolini, G., Catelli, E., Borchert-Stuhlträger, M., Zoller, H., Thomas, E. and Camarda, A. 2020. Vector-borne transmission of Salmonella enterica serovar Gallinarum and fowl typhoid disease mediated by the poultry red mite. Parasit. Vect. 13, 513–522. Davidson, K.E., Byrne, B.A., Pires, A.F.A., Magdesian, K.G. and Pereira, R.V. 2018. Trends in antimicrobial resistance in fecal Salmonella isolates from northern California dairy cattle admitted to a veterinary teaching hospital from 2002 to 2016. PLoS. One 13, 199928; e0199928. Elpers, L., Kretzschmar, J., Nuccio, S., Bäumler, A.J. and Hense, M. 2012. Factors required for the adhesion of S. enterica serovar Typhimurium to corn salad (Valerianella locusta). Appl. Environ. Microbiol. 86, 2757. ElSharkawy, H., Tahoun, A., ElGohary, A.E.A., ElAbasy, M., ElKhayat, F., Gillespie, T., Kitade, Y., Hafez, H.M., Neubauer, H. and ElAdawy, H. 2017. Epidemiological and molecular characterization of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathogens 9, 8–24. Fanissa, F., Effendi, M.H., Tyasningsih, W. and Ugbo, E.N. 2022. Multidrug-resistant Salmonella species isolated from chicken meat sold at Surabaya Traditional Markets, Indonesia. Biodiversitas 23, 2823–2829. Ferrari, R.G., Rosario, D.K.A., Cunha-Neto, A., Mano, S.B., Figueiredo, E.E.S. and Conte-Junior,C.A. 2019. Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl. Environ. Microbiol. 85(14), e00591. Fritz, H.M., Pereira, R.V., Toohey-Kurth, K., Marshall, E., Tucker, J. and Clothier, K.A. 2022. Salmonella enterica serovar dublin from cattle in California from 1993 to 2019: trends in clinical antimicrobial resistance. Antibiotics 11, 1110–1122. Galán-Relaño, A., Valero Díaz, A., Huerta Lorenzo, B., Gómez-Gascón, L., Mena Rodríguez, M.A., Carrasco Jiménez, E., Pérez Rodríguez, F. and Astorga Márquez, R.J. 2023. Salmonella and Salmonellosis: public Health Implications and Control Strategies. Animals 13, 3666–3678. Khairullah, A.R., Rehman, S., Sudjarwo, S.A., Effendi, M.H., Ramandinianto, S.C., Gelolodo, M.A., Widodo, A., Riwu, K.H.P. and Kurniawati, D.A. 2022. Detection of mecA gene and methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and risk factors from farms in Probolinggo, Indonesia. F1000Res. 11(1), 722–735. Khairullah, A.R., Moses, I.B., Yanestria, S.M., Dameanti, F.N.A.E.P., Effendi, M.H., Tang, J.Y.H., Tyasningsih, W., Budiastuti, B., Kusala, M.K.J., Kurniasih, D.A.A., Wardhani, B.W.K., Wibowo, S., Ma’ruf, I.F., Fauziah, I., Ahmad, R.Z. and Latifah, L. 2025. The potential of the livestock industry environment as a reservoir for spreading antimicrobial resistance. Open Vet. J. 15(2), 504–518. Khater, D.F., Lela, R.A., El Diasty, M., Moustafa, S.A. and Wareth, G. 2021. Detection of harmful foodborne pathogens in food samples at the points of sale by MALDT-TOF MS in Egypt. BMC Res. 58, 112: 1-6. Laitinen, K., Kallio, K.A., Niskanen, E.A. and Hirvonen, M.R. 2020. Salmonella in the food supply chain: a comprehensive review of risk factors, control strategies, and their implications on human health. Compreh. Rev. Food Sci. Food Saf. 19(2), 753–777; doi:10.1016/j.compreh.2015.09.020 Lee, K.M., Runyon, M., Herrman, T.J., Phillips, R. and Hsieh, J. 2015. Salmonella detection and identification methods: aspects of rapid emergency response and food safety. Food Control 47, 264–276. Mahvish, M., Sohail, S.J., Muhammad, S., Rasheed, A.F., Haleem, T.M., Muhammad, R.H., Imran, R.M., Imaad, R., Asif, I., Muhammad, S.R., Asim, S., Adeel, H.M., Ahmad, A.F., Abdul, R., Muhammad, Z., Kashif, H., Ali, N.R.H., Akasha, T., Sahar, Y., Kashif, K. and Sajjad, R. 2022. Potential mechanisms of tick-borne virus transmission at the virus-tick interface. Front. Microbiol. 13, 145 McLaren, M.R. and Callahan, B.J. 2020. Pathogen resistance may be the microbiome’sprincipal evolutionary advantage. Philos. Trans. R. Soc. Lond. Biol. Sci. 375, 375. Murugaiyan, J., Walther, B., Stamm, I., Abou-Elnaga, Y., Brueggemann-Schwarze, S., Vincze, S., Wieler, L.H., Lubke-Becker, A., Semmler, T. and Roesler, U. 2024. Species differentiation within the S. intermedius group using a refined MALDI-TOF MS database. Clin. Microbiol. Infect. 20, 1007–1015. New Jersey Department of Health, 2017. Salmonellosis caused by non-typhoidal Salmonella spp.Available via http://localhealthnl.gov Ofori, L.A., Fosu, D., Ofori, S., Akenten, C.W., Flieger, A., Simon, S., Jaeger, A., Lamshöft, M., May, J., Obiri-Danso, K., Phillips, R., Chercos, D.H., Paintsil, E.K. and Dekker, D. 2023. Salmonella enterica in farm environments in the Ashanti Region of Ghana. BMC. Microbiol. 23, 370. Oludairo, O.O., Aiyedun, J.O., Olorunshola, I.D., Bale, J.O.O. and Akintola, O.O. 2020. Physical properties, correlation and regression analyses of potable water in Ilorin, Nigeria. J. Res. Wild. Environ. 12(2), 225–230. Oludairo, O.O., Kwaga, J.K.P., Junaid, K., Paul, A.A., Arya, G., Ann, P., Veronica, C., Antonia, L., Julius, O.A., Oluwafemi, B.D., Isaac, D.O. and Uduak, A. 2023. Salmonella tTransmission in humans and animals and its epidemiological factors. Zagazig Veter. Jour. 51(1), 76–91. Paniel, N. and Noguer, T. 2019. Detection of Salmonella in food matrices using conventional methods and recent aptamer-sensing technologies. Foods 8(9), 371–385. Parolini, F., Ventura, G., Rosignoli, C., Rota Nodari, S., D’Incau, M., Marocchi, L., Santucci, G., Boldini, M. and Gradassi, M. 2024. Detection and Phenotypic Antimicrobial Susceptibility of Salmonella enterica Serotypes in Dairy Cattle Farms in the Po Valley, Northern Italy. Animals 14, 2043–2055. Persad, A.K., Fahmy, H.A., Anderson, N., Cui, J., Topalcengiz, Z., Jeamsripong, S., Spanninger, P.M., Buchanan, R.L., Kniel, K.E., Jay-Russell, M.T., Danyluk, M.D., Rajashekara, G. and Lejeune, J.T. 2022. Identification and subtyping of Salmonella isolates using matrix-assisted laser aesorption–ionization time-of-flight mass spectrometry (MALDI-TOF). Microorganisms 10, 688–702. Pharo, F., Serrenho, R.C., Greer, A.L., Oremush, R., Habing, G., Gillies, M., Keunen, A. and Renaud, D.L. 2025. Exploring the impact and transmission of Salmonella Dublin in crossbred dairy calves. Dairy Sci. 108, 4225–4233. Pradika, A.Y., Chusniati, S., Purnama, M.T.E., Effendi, M.H., Yudhana, A. and Wibawati, P.2019. Total test of Escherichia coli on fresh cow milk at dairy farmer cooperative (KPSP) Karyo Ngremboko Purwoharjo Banyuwangi. J. Med. Vet. 2(1), 1–6. Rafif Khairullah, AAgus Sudjarwo, S., Helmi Effendi, M., Chasyer Ramandinianto, S., Aega Gololodo, M., Hendriana Priscilia Riwu, K. and Ayu Kurniawati, D.F1000Research 11(1), 722–735. Rahman, M.M., Hossain, H., Chowdhury, M.S.R., Hossain, M.M., Saleh, A., Binsuwaidan, R., Noreddin, A., Helmy, Y.A. and El Zowalaty, M.E. 2024. Molecular Characterization of Multidrug-Resistant and Extended-Spectrum β-Lactamases-Producing Salmonella enterica Serovars Enteritidis and Typhimurium Isolated from Raw Meat in Retail Markets. Antibiotics 13, 586–598. Rizzo, D.M., Lichtveld, M., Mazet, J.A.K., Togami, E. and Miller, S.A. 2021. Plant health and its effects on food safety and security in a One Health framework: four case studies. One Health Outlook 3(6), 6–14. Salaheen, S., Sonnier, J., Kim, S.W., Haley, B.J. and Van Kessel, J.A.S. 2020. Interaction of Salmonella enterica with bovine epithelial cells demonstrates serovar-specific association and invasion patterns. Foodborne Dis. 17, 608–615. Shen, X., Zhang, A., Gu, J., Zhao, R., Pan, X., Dai, Y., Yin, L., Zhang, Q., Hu, X., Wang, H. and Zhang, D. 2022. Evaluating Salmonella pullorum dissemination and shedding patterns and antibody production in infected chickens. BMC. Vet. Res. 18, 240–250. Srednik, M.E., Lantz, K., Hicks, J.A., Morningstar-Shaw, B.R., Mackie, T.A. and Schlater, L.K. 2021. Antimicrobial resistance and genomic characterization of isolates of Salmonella Dublin in cattle from the United States. PLoS. One 16(9). Tanner, J., .R., and Kingsley, R and .A. 2018. Salmonella evolution within hosts. Trends Microbiol. 26(12), 986–998. Tarabees, R., Elsayed, M.S.A., Shawish, R., Basiouni, S. and Shehata, A.A. 2017. Isolation and characterization of S. enteritidis and S. typhimurium from chicken meat in Egypt. J. Infect. Dev. Ctries. 11(4), 314–319. Temesgen, L., Tufa, T.B. and Abunna, F. 2025. Isolation, identification, and antimicrobial resistance profile of Salmonella in raw cow milk and its products in Bishoftu, central Ethiopia: implications for public health. One Health Outlook 7(10), 1–12. Topalcengiz, Z., Spanninger, P.M., Jeamsripong, S., Persad, A.K., Buchanan, R.L., Saha, J., Russell, M.T., Kniel, K.E. and Danyluk, M.D. 2020. Survival of Salmonella in various wild animal feces that may contaminate produce. Food Prot. 83, 651–660. Ugbo, E.N., Effendi, M.H., Witaningrum, A.M., Tyasningsih, W., Agumah, B.N., Ugbo, A.I., Nnabugwu, C.C. and Okata-Nwali, D.O. 2023. Antimicrobial resistance pattern of Salmonella spp. isolated from poultry farms in Abakaliki, Nigeria. Biodiversitas 24, 5207–5214. Wang, Y., Xu, X., Jia, S., Qu, M., Pei, Y., Qiu, S., Zhang, J., Liu, Y., Ma, S., Lyu, N., Hu, Y., Li, J., Zhang, E., Wan, B., Zhu, B. and Gao, G.F. 2025. A global atlas and drivers of antimicrobial resistance in Salmonella during 1900-2023. Nat. Commun. 16(4611), 1–17. Wibisono, F.M., Faridah, H.D., Wibisono, F.J., Tyasningsih, W., Effendi, M.H., Witaningrum,A.M. and Ugbo, E.N. 2021. Detection of invA virulence gene of multidrug-resistant Salmonella species isolated from the cloacal swab of broiler chickens in Blitar district, East Java, Indonesia. Vet. World 14(12), 3126–3131. Wibisono, F.M., Wibisono, F.J., Effendi, M.H., Plumeriastuti, H., Hidayatullah, A.R., Hartadi,E.B. and Sofiana, E.D. 2020. Salmonellosis on poultry farms: public health importance. Sys. Rev. Pharm. 11(9), 481–486. Widodo, A., Lamid, M., Effendi, M.H., Khairullah, A.R., Riwu, K.H.P., Yustinasari, L.R., Kurniawan, S.C., Ansori, A.N.M., Silaen, O.S.M. and Dameanti, F.N.A.E.P. 2022. Antibiotic sensitivity profile of multidrug-resistant Escherichia coli isolated from milk of dairy cows in Probolinggo, Indonesia. Biodiversitas 23(10), 4971–4976. World Health Organization. 2018. Critically important antimicrobials for human medicine— 5th Revision 2016—ranking of medically important antimicrobials for risk management of antimicrobial resistance due to non-human use. Geneva, Switzerland: World Health Organization.e-mail [at] e-mail.com World Health Organization. 2022. Multi-country outbreak of S. Typhimurium linked to chocolate products in Europe and the United States Available via https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON369. Yanestria, S.M., Dameanti, F.N.A.E.P., Musayannah, B.G., Pratama, J.W.A., Witaningrum,A.M., Effendi, M.H. and Ugbo, E.N. 2022. Antibiotic resistance pattern of extended- spectrum β-lactamase (ESBL) producing Escherichia coli isolated from a broiler farm in Pasuruan district, Indonesia. Biodiversitas 23(9), 4460–4465. Yanestria, S.M., Rahmaniar, R.P., Wibisono, F.J. and Effendi, M.H. 2019. Detection of invA gene of Salmonella from milkfish (Chanos chanos) at Sidoarjo wet fish market, Indonesia, using polymerase chain reaction technique. Vet. World 12(1), 170–175. Yunita, M.N., Effendi, M.H., Rahmaniar, R.P., Arifah, S. and Yanestria, S.M. 2020. Identification of the spa gene for strain typing of methicillin-resistant Staphylococcus aureus isolated from nasal swabs of dogs. Biochem. Cell. Arch. 20(1), 2999–3004. | ||
| How to Cite this Article |
| Pubmed Style Ugbo EN, Effendi MH, Ugbo AI, Agumah BN, Unegbu VN, Raharjo HM, Tyasningsih WH, Tang JYH, Budiastuti B, Rehman S. Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria. Open Vet. J.. 2025; 15(9): 4393-4402. doi:10.5455/OVJ.2025.v15.i9.46 Web Style Ugbo EN, Effendi MH, Ugbo AI, Agumah BN, Unegbu VN, Raharjo HM, Tyasningsih WH, Tang JYH, Budiastuti B, Rehman S. Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria. https://www.openveterinaryjournal.com/?mno=253373 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i9.46 AMA (American Medical Association) Style Ugbo EN, Effendi MH, Ugbo AI, Agumah BN, Unegbu VN, Raharjo HM, Tyasningsih WH, Tang JYH, Budiastuti B, Rehman S. Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria. Open Vet. J.. 2025; 15(9): 4393-4402. doi:10.5455/OVJ.2025.v15.i9.46 Vancouver/ICMJE Style Ugbo EN, Effendi MH, Ugbo AI, Agumah BN, Unegbu VN, Raharjo HM, Tyasningsih WH, Tang JYH, Budiastuti B, Rehman S. Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria. Open Vet. J.. (2025), [cited January 25, 2026]; 15(9): 4393-4402. doi:10.5455/OVJ.2025.v15.i9.46 Harvard Style Ugbo, E. N., Effendi, . M. H., Ugbo, . A. I., Agumah, . B. N., Unegbu, . V. N., Raharjo, . H. M., Tyasningsih, . W. H., Tang, . J. Y. H., Budiastuti, . B. & Rehman, . S. (2025) Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria. Open Vet. J., 15 (9), 4393-4402. doi:10.5455/OVJ.2025.v15.i9.46 Turabian Style Ugbo, Emmanuel Nnabuike, Mustofa Helmi Effendi, Agatha Ifunanya Ugbo, Bernard Nnabuife Agumah, Valentine Nnachetam Unegbu, Hartanto Mulyo Raharjo, Wiwiek Helmi Tyasningsih, John Yew Huat Tang, Budiastuti Budiastuti, and Saifur Rehman. 2025. Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria. Open Veterinary Journal, 15 (9), 4393-4402. doi:10.5455/OVJ.2025.v15.i9.46 Chicago Style Ugbo, Emmanuel Nnabuike, Mustofa Helmi Effendi, Agatha Ifunanya Ugbo, Bernard Nnabuife Agumah, Valentine Nnachetam Unegbu, Hartanto Mulyo Raharjo, Wiwiek Helmi Tyasningsih, John Yew Huat Tang, Budiastuti Budiastuti, and Saifur Rehman. "Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria." Open Veterinary Journal 15 (2025), 4393-4402. doi:10.5455/OVJ.2025.v15.i9.46 MLA (The Modern Language Association) Style Ugbo, Emmanuel Nnabuike, Mustofa Helmi Effendi, Agatha Ifunanya Ugbo, Bernard Nnabuife Agumah, Valentine Nnachetam Unegbu, Hartanto Mulyo Raharjo, Wiwiek Helmi Tyasningsih, John Yew Huat Tang, Budiastuti Budiastuti, and Saifur Rehman. "Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria." Open Veterinary Journal 15.9 (2025), 4393-4402. Print. doi:10.5455/OVJ.2025.v15.i9.46 APA (American Psychological Association) Style Ugbo, E. N., Effendi, . M. H., Ugbo, . A. I., Agumah, . B. N., Unegbu, . V. N., Raharjo, . H. M., Tyasningsih, . W. H., Tang, . J. Y. H., Budiastuti, . B. & Rehman, . S. (2025) Public health impact of multidrug-resistant Salmonella enterica serovars identified via MALDI-TOF MS in cattle from Abakaliki abattoirs, Nigeria. Open Veterinary Journal, 15 (9), 4393-4402. doi:10.5455/OVJ.2025.v15.i9.46 |