| Research Article | ||
Open Vet. J.. 2025; 15(9): 4520-4526 Open Veterinary Journal, (2025), Vol. 15(9): 4520-4526 Research Article Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi ArabiaBasant Mohammed Sayed, Maryam Bader Bakhashween, Safa Naji Aljumah, Sarah Amin Alduraia, Baraa Falemban and Jamal Hussen*Department of Microbiology, College of Veterinary Medicine, King Faisal University, Al-Ahsa, Saudi Arabia *Corresponding Author: Jamal Hussen. Department of Microbiology, College of Veterinary Medicine, King Faisal University, Al-Ahsa, Saudi Arabia. Email: jhussen [at] kfu.edu.sa Submitted: 13/04/2025 Revised: 24/07/2025 Accepted: 10/08/2025 Published: 30/09/2025 © 2025 Open Veterinary Journal
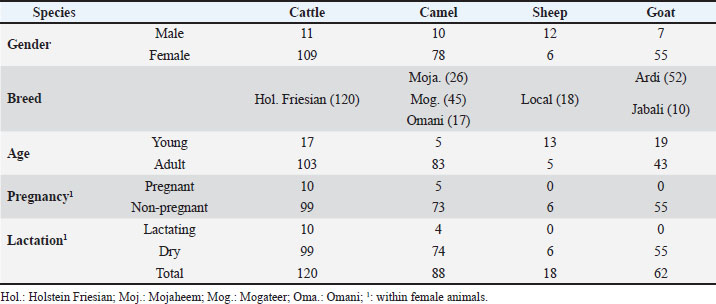
AbstractBackground: Several studies have reported infections with Helicobacter (H) pylori in different veterinary species, raising concerns about the zoonotic potential of this gastric ulcer and cancer-causing pathogen. Aim: The present study was conducted to test the presence of anti-H. pylori antibodies in serum collected from different animal species in the Eastern Region of Saudi Arabia. In addition, the impact of some physiologic factors, including age, gender, breed, and reproduction status of animals, on the prevalence of H. pylori in the livestock species studied was discussed. Methods: This study was conducted on 288 animals kept at different farms in the Eastern Region of Saudi Arabia. The animals included 120 cattle, 88 camels, 18 sheep, and 62 goats. The prevalence of anti-H. pylori antibodies in animal serum samples were investigated using a commercial indirect enzyme-linked immunosorbent assay (ELISA) kit along with a protein A/G conjugate, enabling antibody detection in multiple species. Samples with ODsample /ODcontrol ≥ 1.1 were considered positive. Samples with an ODsample/ ODcontrol ratio ≥ 0.8 to < 1.1 were considered borderline positive. Samples with ODsample/ODcontrol < 0.8 were considered negative. Results: Seroprevalence ratios were calculated as percentages of positive samples related to the total number of samples tested for each species. The results revealed the presence of H. pylori antibodies in serum samples collected from cattle, camel, and goat, with the highest prevalence observed in cattle. For all species, 16 samples were positive for H. pylori with an overall prevalence of 5.6%, 24 samples (8.3%) were considered borderline, and 248 samples (86.1%) were negative. The highest seroprevalence was found in the cattle population (10% seropositive and 15% borderline positive), followed by goats (3.2% seropositive and 3.2% borderline positive) and camels (2.2% seropositive and 4.5% borderline positive). All tested sheep samples were negative for H. pylori antibodies. Conclusion: To the best of our knowledge, this study represents the first report on the prevalence of H. pylori antibodies in serum from different animal species in Saudi Arabia. The results of the current study support the potential role of ruminants as a natural reservoir of H. pylori, highlighting the need for further research to investigate the zoonotic potential of this bacterium and the role of animals in human infections with this pathogen in Saudi Arabia. Keywords: Helicobacter pylori, Cattle, Camel, Seroprevalence, Sheep and goat. IntroductionThe gram-negative bacterium Helicobacter pylori (H. pylori) is an emerging bacterial pathogen that poses a major public health threat (Bianchini et al., 2015; Vouga and Greub, 2016). The bacterium has a high affinity for the stomach mucosa and is considered the main pathogen responsible for gastric ulcers in humans. Additionally, H. pylori has been classified by the International Agency for Research on Cancer as a class I carcinogen due to its association with gastric cancer (Salvatori et al., 2023). Epidemiological studies indicate a high to very high global prevalence of H. pylori in the human population, with infection rates in adults ranging between 35% and 55%. Geographic area, age, race, and ethnicity are among the major factors responsible for the variability in the reported prevalence ratios (Hooi et al., 2017; Chen et al., 2024). Comparative meta-analysis studies have demonstrated a higher global prevalence of H. pylori infection in developing countries than in developed countries (Tibasima et al., 2025). In Saudi Arabia, the prevalence of H. pylori was reported to be between 28% and 46% among adults and approximately 40% among children (Marie, 2008; Akeel et al., 2018; Alkharsah et al., 2022). Although humans represent the main reservoir of H. pylori (Payao and Rasmussen, 2016), the transmission route remains poorly understood. Bacteria can be transmitted to humans and animals through contaminated food and water (Duan et al., 2023). Several studies have reported the zoonotic potential of H. pylori, with some suggesting it as a food-borne pathogen (Rahimi and Kheirabadi, 2012; Mladenova-Hristova et al., 2017; Guessoum et al., 2018). The identification of H. pylori in animal feces, milk, and stomach tissue samples led to the assumption that animals may play a role in disease transmission to humans (Mousavi et al., 2014; Elhariri et al., 2018). Molecular testing of feces and milk samples from healthy sheep, cows, and buffaloes revealed a higher prevalence of H. pylori in cows than in buffaloes and sheep (Elhariri et al., 2018). Sequencing of isolated bacteria revealed the presence of the cagA+vacA+s1a m1 i1 genotype, which is associated with pathogen virulence in humans (Elhariri et al., 2018). Bacterial DNA could also be identified in dairy product samples with the presence of genes encoding several virulence factors (Mousavi et al., 2014). Furthermore, antibiotic-resistant strains that may cause severe infections in humans have been identified in isolated bacteria (Mousavi et al., 2014). Given the high prevalence of H. pylori in Saudi Arabia and the lack of studies on the role of local animals in human infection with the bacterium, the present study was conducted to test the presence of anti-H. pylori antibodies in serum samples collected from different animal species in the Eastern Region of Saudi Arabia. Materials and MethodsAnimalsThe study was conducted in March and April 2025 using previously collected serum samples. A total of 288 animals were included in this study. Animals included 120 cattle, 88 camels, 18 sheep, and 62 goats (Table 1). The cattle, sheep, and goats were housed on the Research Farm (KFU_RF) of King Faisal University (Al-Ahsa, Saudi Arabia). The camels were obtained from the Camel Research Center (CRC) and seven additional private farms in the Eastern Province of Saudi Arabia. The cattle population was of the Holstein Friesian breed and comprised 109 females and 11 males, with 17 young animals (≤ 2 years) and 103 adult (>2 years) animals. Among the female cattle, there were 10 pregnant and lactating cows. The sampled camels included 10 males and 78 females, 5 young (≤ 2 years), and 83 adult camels (> 2 years), of the Mojaheem (n=26 camels), Mogateer (n=45 camels), and Omani camel breeds (n=17 camels). Within the female camels, there were 5 pregnant and 4 lactating she-camels. The sheep group contained 12 male and 6 female animals of a local sheep breed and included 5 young (≤ 6 months) and 13 adult animals (> 6 months). The sampled goat population comprised 7 male and 55 female goats, 19 young (≤ 6 months) and 43 adult animals (> 6 months) of the Ardi (n=52 goats) and Jabali (n=10 goats) breeds. All female sheep and goats were nonpregnant and nonlactating. All sampled animals were clinically healthy based on a trained veterinarian’s clinical examination. Blood samplingFor all animals, 5 ml blood samples were collected from the jugular vein using Vacutainer serum collection tubes (Guangzhou Improve Medical Instruments Co., Ltd., Guangzhou, China) and transported to the laboratory for serum separation within 2 hours of collection. Serum was separated by centrifugation of blood samples at 1,000 ag for 15 minutes at room temperature (RT). Serum samples were stored at −40 °C until analysis. Enzyme-linked immunosorbent assay for the detection of H. pylori antibodiesThe H. pylori IgG ELISA kit (EUROIMMUN Medizinische Labordiagnostika AG, Lübeck, Germany) was used to analyze the seroprevalence of H. pylori as previously described with some modifications (Reynders et al., 2012). The test kit is an indirect enzyme-linked immunosorbent assay test designed for serological detection of IgG antibodies against H. pylori antigens in human serum. Serum samples collected from cattle, camels, goats, and sheep were diluted 1: 100 in sample dilution buffer (included in the kit), and diluted samples (100 µl/well) were added to the wells of the microtiter ELISA plates precoated with the H. pylori antigen (bacterial lysate of H. pylori strain ATCC43504). Control sera with known anti-H. pylori IgG concentrations (200, 20, 2, and 0 RU/ml) were also added to specific wells. After incubation for 30 minutes at RT, the plates were washed three times with 300 µl/well of the washing buffer included in the kit. Subsequently, the wells were filled (100 µl/well) with the horseradish peroxidase (HRP) conjugate of protein A/G (Thermofisher Scientific, Dreieich, Germany) diluted in washing buffer (1:30.000). After incubation for 30 minutes at RT, the plates were washed three times again, and the kit-included TMB substrate (100 µl/well) was added to the wells, followed by incubation in the dark for 30 minutes at RT. Finally, the reaction was stopped by adding the kit-included stop reaction (100 µl/well), and the plate was read on an ELISA reader at 450 nm wavelength. Statistical analysisThe ELISA results were calculated from the OD values using Microsoft Excel. The results were evaluated by calculating the ratio between the OD of the sample (ODsample) and the OD of the positive control (20 RU/ml). Samples with ODsample/ ODcontrol ≥ 1.1 were considered positive. Samples with ODsample/ ODcontrol ratio ≥ 0.8 to < 1.1 were considered borderline positive. Samples with ODsample/ ODcontrol < 0.8 were considered negative. Seroprevalence ratios were calculated as percentages of positive samples related to the total number of samples tested for each species (Khosravi et al., 2021). Table 1. Information about the animals (gender, breed, age, pregnancy, and lactation).
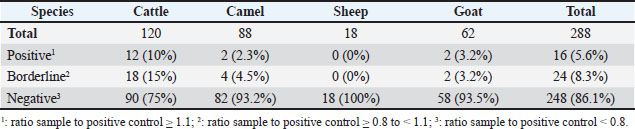
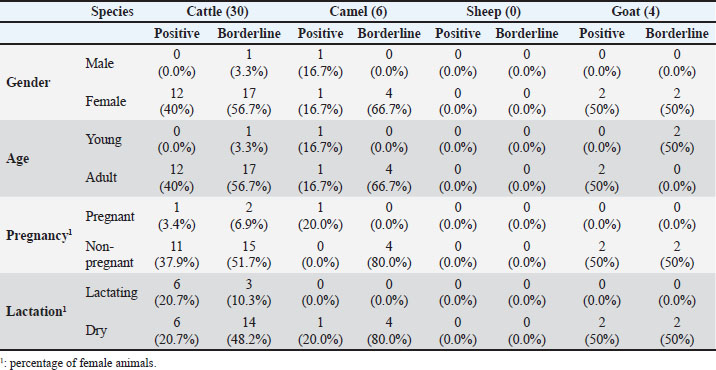
Ethical approvalThis study was conducted in accordance with the principles of the Declaration of Helsinki. The Ethics Committee of King Faisal University, Saudi Arabia, approved this study (KFU-REC-2025-MAR- ETHICS3208). ResultsA total of 288 serum samples were collected from 120 cattle, 88 camels, 18 sheep, and 62 goats kept on different farms in the Al-Ahsa Region in the Eastern Province of Saudi Arabia. The samples were evaluated for the presence of antibodies against the gram-negative bacterium H. pylori. For all species, 16 samples were positive for H. pylori with an overall prevalence of 5.6%, 24 samples (8.3%) were considered borderline, and 248 samples (86.1%) were negative (Table 2). The highest seroprevalence was found within the cattle population, with 12 positive and 18 borderline samples corresponding to 10% seropositivity and 15% borderline positivity, respectively (Table 2). All the positive and borderline cattle were female except for one male. All positive and borderline cattle were adult animals, except for one young animal. The borderline female animals included three lactating and 14 non-lactating cows, and two pregnant and 15 non-pregnant cows. The seropositive female animals included six lactating and six non-lactating cows and one pregnant and 11 non-pregnant cows (Table 3). Among the tested goat samples, two samples were positive for H. pylori antibodies (3.2%) and two were borderline positive (3.2%). All positive goats were female animals of the Ardi breed, including two positive adults and two borderline young goats. All tested sheep samples were negative for H. pylori antibodies. Within the camel population, two animals were found positive (one of the Mogateer and one of the Mojaheem breed), and four animals (two of the Mojaheem and two of the Omani breed) were found borderline for H. pylori antibodies, representing a seropositivity of 2.2% and borderline positivity of 4.5%, respectively. The positive animals (positive and borderline) included five females (all non-lactating and one pregnant) and one male camel. All the camels were adult animals, except for one young animal. DiscussionThe gram-negative spiral bacterium H. pylori is the causative agent of stomach ulcers in humans (Shaaban et al., 2023). Although the exact route of infection remains unknown, transmission through the oral-oral or fecal-oral routes has been proposed (Duan et al., 2023; Costa et al., 2024). The zoonotic potential of H. pylori has been reported in different studies (Rahimi and Kheirabadi, 2012; Mladenova-Hristova et al., 2017; Guessoum et al., 2018). Although a high prevalence of H. pylori was reported in humans from Saudi Arabia, studies on the role of local animals in disease transmission to humans are still limited. Therefore, the present study was conducted to test the presence of anti-H. pylori antibodies in serum samples collected from different animal species in the Eastern Region of Saudi Arabia. The assumption of a zoonotic potential of H. pylori first came from epidemiological studies reporting the higher prevalence of H. pylori infection in professionals involved in animal handling or dealing with animal products, such as abattoir and meat professionals (Morris et al., 1986; Vaira et al., 1988). Subsequently, several studies were conducted in different countries and geographical areas of the world on the presence of H. pylori in animals with different prevalence results related to animal species and other factors (Dore et al., 2001; Turutoglu and Mudul, 2002; Rahimi and Kheirabadi, 2012; Mladenova-Hristova et al., 2017; Elhariri et al.et al., 2018; Guessoum et al.et al., 2018; ). The results of the present study are in line with those of previous studies reporting a higher prevalence of H. pylori in cows than in other ruminants (Elhariri et al., 2018). Similarly, Talaei et al. (2015) reported a higher prevalence of H. pylori in cattle (16%) than in sheep (13.7%), goats (4.7%), and camel (13.3%) (Talaei et al., 2015). Table 2. Prevalence of H. pylori antibodies in serum samples from cattle, camels, sheep, and goats from the Al-Ahsa Region in the Eastern Province of Saudi Arabia.
Table 3. Animals with H. pylori antibodies (positive and borderline) according to species, sex, age, pregnancy status, and lactation status.
Owing to the reported high prevalence of H. pylori in milk and tissue samples of adult sheep, a possible role of sheep as a natural source for H. pylori transmission to humans has been suggested (Dore et al., 2001; Saeidi and Sheikhshahrokh, 2016). However, some researchers have reported no prevalence of H. pylori in sheep. For example, Turutoglu et al. (2002) investigated the presence of H. pylori in 440 raw sheep milk samples collected from the Burdur region of Turkey and found that none of the samples were positive for H. pylori culture (Turutoglu and Mudul, 2002). In the present study, whether the lack of positive samples among tested sheep is due to the limited age range of animals involved in the study, with most animals being 2 months old, is to be elucidated in future studies involving sheep with a wider age range. Several predisposing factors for H. pylori infection in humans and animals have been described in the literature (Shaaban et al., 2023). In a recent study from Egypt, H. pylori prevalence in humans was affected by sex, age, and locality. The same study reported a higher frequency of H. pylori infection in pet animals than in farm animals (Shaaban et al., 2023). Nutritional and environmental factors such as concentrated feed formulations, disruption in feed delivery, and increased environmental temperature are additional predisposing factors reported in animals (Taillieu et al., 2022). Although the present study evaluated the impact of some physiologic factors, including age, gender, breed, and reproduction status (pregnancy and lactation), on the prevalence of H. pylori in the studied livestock species, the low overall prevalence of the antibodies with the limited number of animals in each category make it difficult to conclude the role of these factors as risk factors for H. pylori infections. This highlights the need for further studies on higher numbers of animals with comparable representation of gender, age, breed, and reproduction status category. Several studies have reported the presence of H. pylori in camels, with prevalence ratios ranging between 3.5% and 13% (Rahimi and Kheirabadi, 2012; Ranjbar et al., 2018; Quaglia and Dambrosio, 2018). In a recent report, Elbir et al. (2019) identified the DNA of the H. pylori bacterium in 30% of Hyalomma dromedarii tick samples collected from camels in the Eastern Province of Saudi Arabia, indicating a potential role of camels as a reservoir for this human pathogen (Elbir et al., 2019). Several authors used the ELISA technique for seroprevalence analysis of H. pylori in humans and animals. For example, Guessoum et al. used a commercial indirect ELISA kit for the detection of anti-H. pylori antibodies in serum and milk samples collected from Algerian cattle and found 12% and 4% prevalence rates in serum and milk, respectively (Guessoum et al., 2018). The same study revealed high alignment between IgG detection using ELISA and molecular detection of H. pylori genes using PCR (Guessoum et al., 2018). A multispecies indirect ELISA test was used to test serum samples from several animal species for H. pylori antibodies. To detect H. pylori-specific antibodies, a mixture of staphylococcal protein A and protein G conjugated to HRP was used as a conjugate. Due to their species-specific binding affinity to the different IgG isotypes (Sheng and Kong, 2012), using a combination of both protein A and protein G would support the detection of all IgG isotypes from the different species. However, the low affinity of these bacterial proteins to antibodies of the IgM isotype could result in the lack of sensitivity of this ELISA test to H. pylori-specific IgM in animals with recent infection. In addition, whether using species-specific conjugated detection antibodies instead of proteins A and G would improve the sensitivity of the ELISA test should be investigated. In addition, combined serological, bacteriological, and molecular studies are required to identify the link between the presence of H. pylori in animal tissue or secretions and the detection of H. pylori antibodies in serum. Limitations of the studyAlthough several animal species were included in the present study, the lack of human subjects makes it difficult to draw a conclusion about the zoonotic potential of the bacterium. The current study is limited by the low number of studied animals in each species, making it difficult to identify the role of age, gender, breed, and reproduction status as risk factors for H. pylori infections in the studied livestock species. In addition, the use of antibody detection methods to calculate the prevalence of H. pylori, with the lack of bacteriological or molecular methods to identify the agent, represents a limitation of the study. Conclusions and RecommendationsThe current study represents the first report on the seroprevalence of Helicobacter pylori in different ruminant species in the Eastern Province of Saudi Arabia. The results revealed the presence of H. pylori antibodies in serum samples collected from cattle, camel, and goat, with the highest prevalence observed in cattle. These results support the potential role of ruminants as a natural reservoir of Helicobacter pylori, highlighting the need for further research to investigate the zoonotic potential of this bacterium and the role of animals in human infections with this pathogen in Saudi Arabia. Therefore, future studies including animal and human subjects from the same area are recommended. Additionally, microbiological and molecular (PCR) methods targeting antigen detection are essential to confirm seroprevalence results. Furthermore, the targeted geographical area may be extended in future studies to include more provinces in Saudi Arabia to identify the bacterium’s epidimeological status. AcknowledgmentsThe authors would like to thank the Research Farm and the Camel Research Center of King Faisal University (Al-Ahsa, Saudi Arabia) for providing some experimental animals. FundingThis study was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia [KFU252395]. Authors’ contributionsJamal Hussen designed the study, performed data analysis, and wrote the first draft of the manuscript. Baraa Falemban collected the samples, and Basant Mohammed Sayed, Maryam Bader Bakhashween, Safa Naji Aljumah, and Sarah Amin Alduraia tested the samples using enzyme-linked immunosorbent assay. All authors have read and approved the final version of the manuscript. Conflict of interestThe authors have no relevant financial or non-financial interests to disclose. Data availabilityThe datasets generated during the current study are available upon reasonable request from the corresponding author. ReferencesAkeel, M., Elmakki, E., Shehata, A., Elhafey, A., Aboshouk, T., Ageely, H. and Mahfouz, M.S. 2018. Prevalence and factors associated with H. pylori infection in Saudi patients with dyspepsia. Electron. Phys. 10(9), 7279–7286. Alkharsah, K.R., Aljindan, R.Y., Alamri, A.M., Alomar, A.I. and Al-Quorain, A.A. 2022. Molecular characterization of Helicobacter pylori clinical isolates from Eastern Saudi Arabia. Saudi Med. J. 43(10), 1128–1135. Bianchini, V., Recordati, C., Borella, L., Gualdi, V., Scanziani, E., Selvatico, E. and Luini, M. 2015. Helicobacteraceae in bulk tank milk of dairy herds from northern Italy. Biomed. Res. Int. 2015, 639521. Chen, Y.C., Malfertheiner, P., Yu, H.T., Kuo, C.L., Chang, Y.Y., Meng, F.T., Wu, Y.X., Hsiao, J.L., Chen, M.J., Lin, K.P. 2024. Global prevalence of Helicobacter pylori infection and incidence of gastric cancer between 1980 and 2022. Gastroenterology 166(4), 605–619. Costa, L.C.M.C., Carvalho, M.D.G., Vale, F.F., Marques, A.T., Rasmussen, L.T., Chen, T. and Barros-Pinheiro, M. 2024. Helicobacter pylori in oral cavity: current knowledge. Clin. Exp. Med. 24(1), 209. Dore, M.P., Sepulveda, A.R., El-Zimaity, H., Yamaoka, Y., Osato, M.S., Mototsugu, K., Nieddu, A.M., Realdi, G. and Graham, D.Y. 2001. Isolation of Helicobacter pylori from sheep—implications for transmission to humans. Am. J. Gastroenterol. 96(5), 1396–1401. Duan, M., Li, Y., Liu, J., Zhang, W., Dong, Y., Han, Z., Wan, M., Lin, M., Lin, B., Kong, Q., Ding, Y., Yang, X., Zuo, X. and Li, Y. 2023. Transmission routes and patterns of Helicobacter pylori. Helicobacter 28(1), 12945. Elbir, H., Almathen, F. and Alhumam, N.A. 2019. A glimpse of the bacteriome of Hyalomma dromedarii ticks infesting camels reveals human Helicobacter pylori pathogen. J. Infect. Dev. Ctries. 13(11), 1001–1012. Elhariri, M., Hamza, D., Elhelw, R. and Hamza, E. 2018. Occurrence of cagA(+), vacAs1a, mIs1 iIs1 Helicobacter pylori in farm animals in Egypt and ability to survive in experimentally contaminated UHT milk. Sci. Rep. 8(1), 14260. Guessoum, M., Guechi, Z. and Adnane, M. 2018. First-time serological and molecular detection of Helicobacter pylori in milk from Algerian local-breed cows. Vet. World 11(9), 1326–1330. Hooi, J.K.Y., Lai, W.Y., Ng, W.K., Suen, M.M.Y., Underwood, F.E., Tanyingoh, D., Malfertheiner, P., Graham, D.Y., Wong, V.W.S., Wu, J.C.Y. , Chan, F.K.L., Sung, J.J.Y., Kaplan, G.G. and Ng, S.C. 2017. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153(2), 420–429. Khosravi, A.D., Sirous, M., Saki, M., Seyed-Mohammadi, S., Modares Mousavi, S.R., Veisi, H. and Abbasinezhad Poor, A. 2021. Associations between seroprevalence of Helicobacter pylori and ABO/rhesus blood group antigens in healthy blood donors in southwest Iran. J. Int. Med. Res. 49(12), 3000605211058870. Marie, M.A. 2008. Seroprevalence of Helicobacter pylori infection in large series of patients in an urban area of Saudi Arabia. Korean J. Gastroenterol. 52(4), 226–229. Mladenova-Hristova, I., Grekova, O. and Patel, A. 2017. Zoonotic potential of Helicobacter spp. J. Microbiol. Immunol. Infect. 50(3), 265–269. Morris, A., Nicholson, G., Lloyd, G., Haines, D., Rogers, A. and Taylor, D. 1986. Seroepidemiology of Campylobacter pyloridis. N. Z. Med. J. 99(809), 657–659. Mousavi, S., Dehkordi, F.S. and Rahimi, E. 2014. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. J. Venom. Anim. Toxins Incl. Trop. Dis. 20, 51. Payão, S.L. and Rasmussen, L.T. 2016. Helicobacter pylori and its reservoirs: a correlation with the gastric infection. World J. Gastrointest. Pharmacol. Ther. 7(1), 126–132. Quaglia, N.C. and Dambrosio, A. 2018. Helicobacter pylori: a foodborne pathogen?. World J. Gastroenterol. 24(31), 3472–3487. Rahimi, E. and Kheirabadi, E.K. 2012. Detection of Helicobacter pylori in bovine, buffalo, camel, ovine, and caprine milk in Iran. Foodborne Pathog. Dis. 9(5), 453–456. Ranjbar, R., Farsani, F.Y. and Dehkordi, F.S. 2018. Phenotypic analysis of antibiotic resistance and genotypic study of the vacA, cagA, iceA, oipA and babA genotypes of the Helicobacter pylori strains isolated from raw milk. Antimicrob. Resist. Infect. Control 7, 115. Reynders, M.B., Miendje Deyi, V.Y., Dahma, H., Scheper, T., Hanke, M., Decolvenaer, M. and Dediste, A. 2012. Performance of individual Helicobacter pylori antigens in the immunoblot-based detection of H. pylori infection. FEMS. Immunol. Med. Microbiol. 64(3), 352–363. Saeidi, E. and Sheikhshahrokh, A. 2016. Vaca genotype status of Helicobacter pylori isolated from foods with animal origin. Biomed. Res. Int. 2016, 8701067. Salvatori, S., Marafini, I., Laudisi, F., Monteleone, G. and Stolfi, C. 2023. Helicobacter pylori and gastric cancer: pathogenetic mechanisms. Int. J. Mol. Sci. 24(3), 2895. Shaaban, S.I., Talat, D., Khatab, S.A., Nossair, M.A., Ayoub, M.A., Ewida, R.M. and Diab, M.S. 2023. An investigative study on the zoonotic potential of Helicobacter pylori. BMC Vet. Res. 19(1), 16. Sheng, S. and Kong, F. 2012. Separation of antigens and antibodies by immunoaffinity chromatography. Pharm. Biol. 50(8), 1038–1044. Taillieu, E., Chiers, K., Amorim, I., Gärtner, F., Maes, D., Van Steenkiste, C. and Haesebrouck, F. 2022. Gastric Helicobacter species associated with dogs, cats and pigs: significance for public and animal health. Vet. Res. 53(1), 42. Talaei, R., Souod, N., Momtaz, H. and Dabiri, H. 2015. Milk of livestock as a possible transmission route of Helicobacter pylori infection. Gastroenterol. Hepatol. Bed. Bench 8(Suppl 1), S30–S36. Tibasima, E.B., Kumbakulu, P.K., Chirac, L.P., Ramazani, O., Patrick, T.B., Olga, K.K., Shamavu, G.K., Prudence, M.N. and Mseza, B. 2025. Prevalence, associated factors, and clinical outcomes of Helicobacter pylori infection in pediatric populations in a war-torn urban environment in Eastern Democratic Republic of Congo: a mixed methods study. BMC. Pediatrics 25(1), 257. Turutoglu, H. and Mudul, S. 2002. Investigation of Helicobacter pylori in raw sheep milk samples. J. Vet. Med. B. Infect. Dis. Vet. Public Health 49(6), 308–309. Vaira, D., D’Anastasio, C., Holton, J., Dowsett, J.F., Londei, M., Bertoni, F., Beltrandi, E., Grauenfels, P., Salmon, P.R. and Gandolfi, L. 1988. Campylobacter pylori in abattoir workers: is it a zoonosis?. Lancet 2(8613), 725–726. Vouga, M. and Greub, G. 2016. Emerging bacterial pathogens: the past and beyond. Clin. Microbiol. Infect. 22(1), 12–21. | ||
| How to Cite this Article |
| Pubmed Style Sayed BM, Bakhashween MB, Aljumah SN, Alduraia SA, Falemban B, Hussen J. Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia. Open Vet. J.. 2025; 15(9): 4520-4526. doi:10.5455/OVJ.2025.v15.i9.57 Web Style Sayed BM, Bakhashween MB, Aljumah SN, Alduraia SA, Falemban B, Hussen J. Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia. https://www.openveterinaryjournal.com/?mno=252402 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i9.57 AMA (American Medical Association) Style Sayed BM, Bakhashween MB, Aljumah SN, Alduraia SA, Falemban B, Hussen J. Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia. Open Vet. J.. 2025; 15(9): 4520-4526. doi:10.5455/OVJ.2025.v15.i9.57 Vancouver/ICMJE Style Sayed BM, Bakhashween MB, Aljumah SN, Alduraia SA, Falemban B, Hussen J. Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia. Open Vet. J.. (2025), [cited January 25, 2026]; 15(9): 4520-4526. doi:10.5455/OVJ.2025.v15.i9.57 Harvard Style Sayed, B. M., Bakhashween, . M. B., Aljumah, . S. N., Alduraia, . S. A., Falemban, . B. & Hussen, . J. (2025) Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia. Open Vet. J., 15 (9), 4520-4526. doi:10.5455/OVJ.2025.v15.i9.57 Turabian Style Sayed, Basant Mohammed, Maryam Bader Bakhashween, Safa Naji Aljumah, Sarah Amin Alduraia, Baraa Falemban, and Jamal Hussen. 2025. Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia. Open Veterinary Journal, 15 (9), 4520-4526. doi:10.5455/OVJ.2025.v15.i9.57 Chicago Style Sayed, Basant Mohammed, Maryam Bader Bakhashween, Safa Naji Aljumah, Sarah Amin Alduraia, Baraa Falemban, and Jamal Hussen. "Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia." Open Veterinary Journal 15 (2025), 4520-4526. doi:10.5455/OVJ.2025.v15.i9.57 MLA (The Modern Language Association) Style Sayed, Basant Mohammed, Maryam Bader Bakhashween, Safa Naji Aljumah, Sarah Amin Alduraia, Baraa Falemban, and Jamal Hussen. "Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia." Open Veterinary Journal 15.9 (2025), 4520-4526. Print. doi:10.5455/OVJ.2025.v15.i9.57 APA (American Psychological Association) Style Sayed, B. M., Bakhashween, . M. B., Aljumah, . S. N., Alduraia, . S. A., Falemban, . B. & Hussen, . J. (2025) Detection of H. pylori antibodies in serum from different animal species in the Eastern Province of Saudi Arabia. Open Veterinary Journal, 15 (9), 4520-4526. doi:10.5455/OVJ.2025.v15.i9.57 |