| Research Article | ||
Open Vet. J.. 2025; 15(9): 4527-4532 Open Veterinary Journal, (2025), Vol. 15(9): 4162-4532 Research Article Morpho-molecular characterization of Trypanosoma evansi in sheepAzhar Ali Faraj, Ali Issa Fadhil* and Howaida Hamel AbedDepartment of Parasitology, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq *Corresponding Author: Ali Issa Fadhil, Department of Parasitology, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq. Email: ali.fadel [at] covm.uobaghdad.edu.iq Submitted: 09/04/2025 Revised: 14/07/2025 Accepted: 05/08/2025 Published: 30/09/2025 © 2025 Open Veterinary Journal
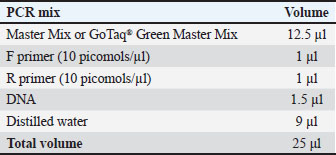
ABSTRACTBackground: Trypanosoma evansi is a protozoan parasite that causes trypanosomiasis, referred to as ‟surra.” It affects a wide variety of both wild and domesticated species on many continents. The primary host species differ geographically; however, camelids, equine, buffalo, and farm animals are at risk of infection. In vector-borne sickness, numerous species of blood-consuming flies, along with Tabanids and Stomoxys, are involved in transporting pathogens from one animal to another, acting as mechanical vectors. Aim: This study was established to evaluate the prevalence of T. evansi in sheep in Baghdad and investigate the impact of age and sex on the infection rate. Methods: A total of 200 blood samples were obtained from October 2023 to March 2024. These samples were examined using Giemsa stain under a light microscope, and 40 positive samples were selected for further investigation using conventional polymerase chain reaction (PCR). Results: The results showed an infection rate of 20%, with significant differences observed between male and female sheep. Younger sheep were found to be significantly more affected than older ones. Ten PCR-detected samples were randomly selected for DNA molecular analysis to obtain ITS-1 gene nucleotide sequences. The PCR product exhibited a band size of 1,264 bp, and the sequences were deposited in the National Center for Biotechnology Information under the following code numbers: PP930358.1, PP930353.1, PP930350.1, PP930357.1, PP930356.1, PP930351.1, PP930359.1, PP930352.1, PP930354.1, and PP930355.1. Conclusion: The current study results indicate that T. evansi occurs and circulates in sheep and confirm that the molecular approach for detecting DNA of Trypanosoma species by the use of ITS1 makes it a highly dependable assay for species recognition of this parasite. Keywords: Trypanosomiasis, Surra, Stable fly, Ovine, PCR. IntroductionTrypanosoma evansi is a protozoan salivarian parasite, classified under the Trypanosomatidae family, and the causative agent of surra disease, also known as trypanosomiasis, in vertebrate animals. Trypanosoma evansi parasitizes a wide range of both wild and domestic animals, such as sheep, horses, camels, cattle, and numerous wildlife animals. The transmission of T. evansi appears to be mechanical and facilitated by stable insects of the genus Stomoxys and horseflies of the species Tabanus (Rjeibi et al., 2015); however, infected animal blood can also be a source of infection through wound contamination (Desquesnes et al., 2013). The pathogenicity of trypanosomiasis depends on many factors, including factors that are related to the parasite, such as the virulence of the strain, and other factors related to the host (nutritional situation, immunity, species, age), and other environmental factors. Severe clinical manifestations of trypanosomiasis, fever, and anemia, combined with other systemic conditions, can lead to death through congestive heart failure. Lymph nodes, along with spleen enlargement, weight loss, pyrexia, ataxia, lethargy, and dropping in animals’ production (Taylor and Authié, 2004), have been associated with many degenerative inflammatory effects, leading to damage to many systems, such as the pituitary gland, eyes, gonads, and central nervous system (Van den Bossche and Delespaux, 2011; Ahmed et al., 2020). Many diagnostic methods, such as microscopic examination of different blood smears and serological and molecular assays, can be performed for blood parasites (Fadhil et al., 2021; Abed et al., 2023). Although blood detection techniques, such as direct smear or serological assays, have been used for many years for T. evansi diagnosis, they have shown low sensitivity in detecting and differentiating various Trypanosoma species in different animals. Thus, molecular assays have been used to overcome the limitations of these methods (Al-Fatlawi and Al-Fatlawi, 2019; Barghash, 2020; Mamman et al., 2021) and improve the diagnosis of the disease in many infections (Sivajothi et al., 2016; Mulenga et al., 2021). Additionally, it was found that polymerase chain reaction (PCR) showed more sensitivity and specificity and was able to detect T. evansi at tiny levels (Desquesnes et al., 2022). Our study was conducted to characterize Trypanosoma in sheep in Baghdad city because of data limitations. Materials and MethodsEthical approvalThe project was approved by the official committee of the College of Veterinary Medicine for Animal Care at the University of Baghdad (approval number: 569). Blood samplesThe time frame for blood sample collection is from the beginning of October 2023 to the end of March 2024. A total of 200 direct blood samples were taken directly from the jugular vein of sheep of various ages and sexes. Anticoagulant ethylene diamine-tetraacetic acid tubes were used to overcome blood coagulation. All samples were preserved and transferred to the Laboratory of the College of Veterinary Medicine, University of Baghdad. All samples were examined under a microscope, and blood smears were stained with Giemsa (Faraj et al., 2019; Abdullah and Ali, 2021). Identified samples were kept at −20°C for further PCR assay. Blood genomic DNA extractionTrypanosoma DNA was extracted from Giemsa-identified samples using the gSYAN DNA mini extraction kit (Frozen Blood) from Geneaid company, USA. The extraction procedure followed the manufacturer’s recommendations. Genomic DNA purity estimationThe genomic DNA extracted was estimated using a Nanodrop (Thermo, USA). The DNA concentration was between 5 and 50 ng/μl, and the absorbance at 260 and 280 nm was used to measure the DNA purity. Polymerase chain reactionFor the detection of Trypanosoma spp., a conventional PCR assay was used using ITS1 gene couple primers, as forward primers: 5’ GGTGATCGGACCGTCGCTCGTCT - 3’ and reverse primer: 5’ CCTCTTCGCTCGCCGCTGACTG - 3’with a PCR product size (1,264 bp) from sheep blood extracted DNA samples (Khuchareontaworn et al., 2007). Primer preparationFor the preparation of the working primer concentration, the primers were lyophilized using ddH2O to give a final needed concentration of 100 pmol/µl to use as stock and kept for the procedure. Reaction setup and thermal cycling protocolPCR master mix preparationConventional PCR master mix was carried out by (AccuPower PCR PreMix Kit), according to the manufacturer’s instructions (Table 1). Subsequently, the PCR master mix constituents were transferred into a thermocycler (T100 Thermal cycler, BioRad, USA). Thermocycler conditionsA PCR thermocycler was used under the conditions listed in Table 2 (T100 Thermal cycler, BioRad. USA). PCR product analysisThe PCR amplicons (1,264 bp) were analyzed by electrophoresis using a 1.5% agarose gel that was melted in a water bath for 15 minutes at 100°C and then left to cool to 50°C. Then, an electric current of 15 V/cm2 was applied for 1:30 hours. Stained with red safe and investigated under a UV transilluminator. DNA sequencing methodPCR amplification showed 10 samples positive for Trypanosoma species from sheep; these 10 positive samples were sequenced. Positive ITS1 gene products were sent to Macrogen Company in Korea for DNA sequencing and packed in an ice bag. For the DNA sequencing analysis, Molecular Evolutionary Genetics Analysis version 7.0 (Mega 7.0) software was used (Kumar et al., 2016), and the ITS1 gene was used for Clustal W alignment analysis and phylogenetic tree UPGMA method using the maximum composite likelihood to calculate divergence distances for the multi-sequence alignment analysis. Statistical analysisAll obtained data were analyzed using SAS version 9.4 (SAS, 2012) using chi-square tests. ResultsMicroscopic detectionUnder the microscope, the morphology of the identified Trypanosoma between infected sheep blood cells was morphologically similar to that of T. evansi, which appeared slender with tapered ends, a relatively long, free flagellum, and a remarkably developed undulating membrane. Moreover, a tiny, spherical, deeply stained kinetoplast that is almost sub-terminal and a centrally situated nucleus were seen (Table 3). Table 1. Volume of PCR reaction master mix contents.
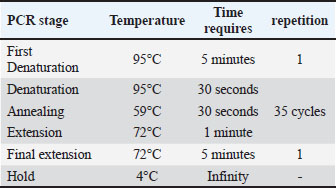
Table 2.Thermocycler required detection conditions.
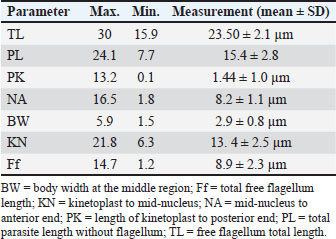
Table 3.Biometric measurements of T. evansi from sheep.
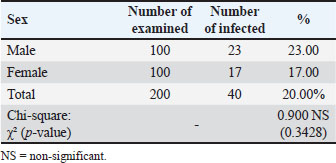
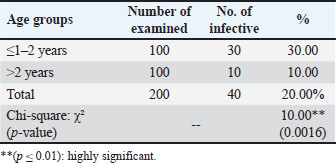
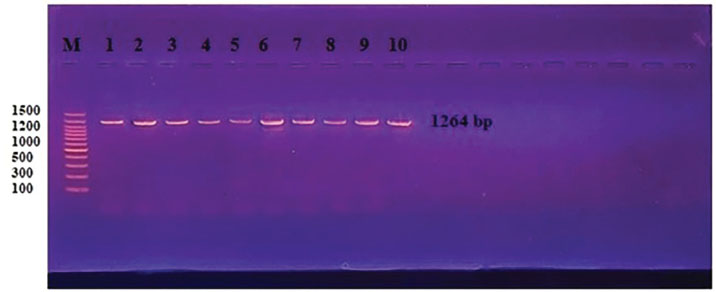
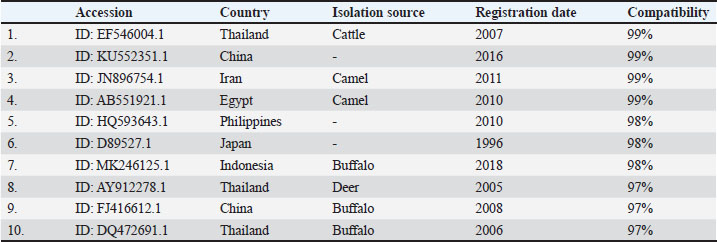
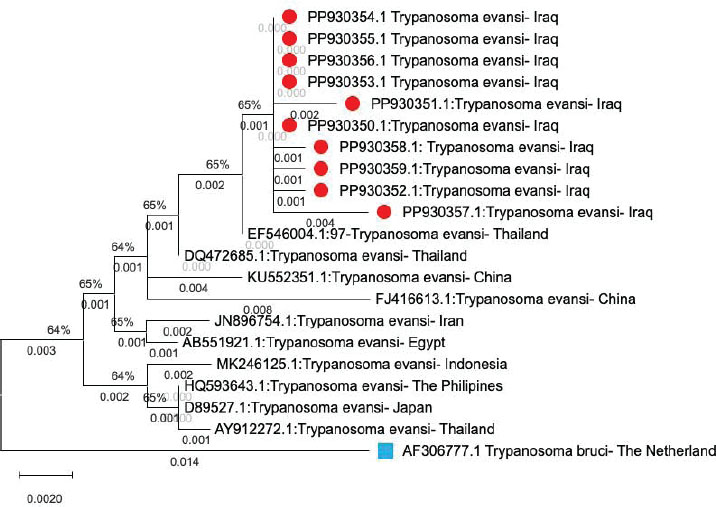
Examination of blood smears previously stained with Giemsa showed that the infection rate of Trypanosoma spp. was 20% (40/200 sheep). Infection rates were 23% in males and 17% in females, without significant differences (p ≤ 0.01) between sexes (Table 4). The results revealed that the highest rate of infection (30%) appeared in the young age group (1–2 years). Followed by (10%) was observed in older sheep (>2 years), with a highly significant difference in Trypanosoma spp. infection (p ≤ 0.01) (Table 5). Molecular diagnosisAll 40 positive blood samples were subjected to PCR, which was performed with microscopic findings and revealed an expected PCR amplicon of 1,264 bp (Fig. 1). In total, 10 PCR amplicons were sequenced and registered in GeneBank with accession numbers as follows: PP930358.1, PP930353.1, PP930350.1, PP930357.1, PP930356.1, PP930351.1, PP930359.1, PP930352.1, PP930354.1, and PP930355. Phytogenic analysisSequencing results revealed that all registered isolates belonging to T. evansi were similar to the previously listed GenBank T. evansi isolates (Table 6 and Fig. 2). Table 4.Rate of infection by T. evansi in sheep according to the sex of the animal.
Table 5.Total rate of infection by T. evansi in sheep based on age.
Fig. 1. PCR analysis of the ITS1 gene of Trypanosome spp. Blood samples show a 1,264-bp band size. The product was electrophoresed on 1.5% agarose gel at 5 V/cm2. 1× TBE buffer for 1:30 hours. M: DNA ladder (100). Table 6. National Center for Biotechnology Information-BLAST nucleotide sequence similarity ratio from recent local T. evansi isolates of sheep compared with previously submitted strains.
Fig. 2. Phylogenetic tree of T. evansi compared with its origin. Red triangle: T. evansi isolated from sheep in Iraq. Blue square: out-group. DiscussionTen thin blood smears stained with Giemsa from each infected sheep with 10–15 microscopic fields per slide, and minimally five trypanosomes were selected for the morphometric measurements. These biometric data were consistent with previous morphometric descriptions of T. evansi (Soulsby, 1982), and these morphometric descriptions may interfere with other trypanosomes. Thus, further techniques may be required for species identification. The outcome of the present investigation revealed that the infection rate of Trypanosoma spp. in sheep was 20%, which aligns with previous studies of Trypanosoma infection in sheep (Mahmood and Al-Obaidii, 2022) when they recorded 21% in Mosul city of Iraq (Bedaso et al., 2017) 25% in Brazil, and 25.3% in Kenya (Musa et al., 2005), but they come conflicts with Hasan (2012) in Mosul city, Tariq et al. (2024) in Pakistan, and Castro-Rodriguez et al. (2015) in Nicaragua recorded 3.3%, 18.75%, and 47%, respectively. Sheep’s trypanosomiasis regarding animals’ sex showed a higher infection rate in males than in females. This result was in contrast with that of Mahmood and Al-Obaidii (2022), who reported that female sheep were more susceptible than male sheep, and this may be related to the sample size and detection methods used. Younger sheep exhibited higher infection rates than adults, which contrasts with the findings of Fasanmi et al. (2014), who expressed that adults were more susceptible to Trypanosoma infection than younger animals. This may reflect the immature immune system of younger animals. It is not yet completely developed to resist infection. The ITS1 spacers have been found to be one of the most reliable in discrete phylogenetic separation of closely related species, including many blood protozoan parasites, and have been used in trypanosome molecular identification. The results of the current study also highlight that T. evansi occurs and circulates in sheep and confirm that the molecular approach for detecting DNA of Trypanosoma species using ITS1 is highly reliable for species recognition of this parasite. Many previous molecular assay studies have validated the use of genetic markers for Trypanosoma detection (Hassan Kadle et al., 2019; Mahmood and Al-Obaidii, 2022). Salim et al. (2011) and Sadek et al. (2021) found that PCR demonstrates higher sensitivity/specificity than serological tests. ConclusionOur findings confirmed sheep infection with T. evansi by direct blood smears and molecular PCR as well as the effect of age and sex factors on the infection rate. These results show the need for further investigation on this parasite and methods for its control. AcknowledgmentThe authors are grateful to the College of Veterinary Medicine of the University of Baghdad. ReferencesAbdullah, S.H. and Ali, S.A. 2021. Molecular study and phylogeny of Babesia spp. in native sheep from Sulaimani governorate/Northern Iraq. Iraqi J. Agricult. Sci. 52(5), 1077–1083; doi: 10.36103/ijas.v52i5.1445 Abed, H.H., Fadhil, A.I., Alhaboubi, A.R. and Faraj, A.A. 2023. Genetic confirmation for morphological identification of Stilesia globipunctata in camels in Iraq. Iraqi J. Vet. Sci. 37(3), 719–724; doi:10.33899/ijvs.2023.137262.2664 Ahmed, A.H., Abdalla M.I., Hamisi, S.N., Abdulkarim, A.Y. and Rafael F.C.V. 2020. Parasitological and molecular detection of Trypanosoma spp. in cattle, goats, and sheep in Somalia. Parasitology 3(1), 322–325; doi: 10.1017/S003118202000178X Al-Fatlawi, M.S.H. and Al-Fatlawi, M.A.A. 2019. Molecular and phylogenetic study of Theileria spp. isolated from ticks in Al Diwaniyah city, Iraq. Iraqi J. Agricult. Sci. 50(1), 574; doi:10.36103/ijas.v50i1.313 Barghash, S.M. 2020. Molecular changes in Trypanosoma evansi after treatment against trypanosomosis. Ann. Parasitol. 66(2), 165–174; doi:10.17420/ap6602.251 Bedaso, K., Seifu, H. and Getachew, T. 2017. Prevalence, and pathogenic significance of trypanosomosis on sheep and goats of Mareka district, Dawro zone, Southwestern Ethiopia. J. Anim. Sci. Livestock Prod. 1, 102. Castro-Rodriguez, K.A. and Mora-Sánchez, B.M. 2015. Infección por Trypanosoma sp. en ovinos sintomáticos en el Municipio de León, Nicaragua. Rev Ci UNAN León. 6(1), 1–10. Desquesnes, M., Gonzatti, M., Sazmand, A., Thévenon, S., Bossard, G., Boulangé, A., Gimonneau, G., Truc, P., Herder, S., Ravel, S. and Sereno, D. 2022. A review on the diagnosis of animal trypanosomoses. Parasites Vectors 15, 64; doi:10.1186/s13071-022-05190-1 Desquesnes, M., Holzmuller, P., Lai, D.H., Dargantes, A., Lun, Z.R. and Jittaplapong, S. 2013. Trypanosoma evansiand surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. BioMed. Res. Int. 2013(1), 194176. Fadhil, A.I., Fadel, S.R. and Faraj, A.A. 2021. Conventional and molecular study of Entamoeba spp. in domestic dogs’ faeces at Baghdad city, Iraq. Iraqi J. Agricult. Sci. 52(6), 1482–1488; doi:10.36103/ijas.v52i6.1489 Faraj, A.A., Hade, B.F. and Al-Amery, A.M. 2019. Conventional and molecular study of Babesia spp. of natural infection in dragging horses at some areas of Baghdad city, Iraq. Iraqi J. Agricult. Sci. 50(3), 909–915. Fasanmi, O.G., Okoroafor, U.P., Nwufoh, O.C., Bukola-Oladele, O.M. and Ajibola, E.S. 2014. Survey for trypanosomiasis species in cattle from three farms in Ido Local Government Area, Oyo State. So koto J. Vet. Sci. 12(1), 57–61; doi:10.4314/sokjvs.v12i1.9 Hasan, M.H. 2012. Diagnosis of some blood parasites in cattle and sheep in Mosul, Iraq. Iraqi J. Vet. Sci. 26, 57–61. Hassan Kadle, A.A., Ibrahim, A.M., Nyingilili, H.S., Yusuf, A.A., Vieira, T.S.W.J. and Vieira, R.F.C. 2019. Parasitological, serological, and molecular survey of Camel Trypanosomiasis in Somalia. Parasites Vectors 12, 598; doi:10.33899/ijvs.2012.168714 Hassan-Kadle, A.A., Ibrahim, A.M., Nyingilili, H.S., Yusuf, A.A. and Vieira, R.F.C. 2020. Parasitological and molecular detection of Trypanosoma spp. in cattle, goats and sheep in Somalia. Parasitology 147(1), 1786–1791; doi: 10.1017/S003118202000178X Khuchareontaworn, S., Singhaphan, P., Viseshakul, N. and Chansiri, K. 2007. Genetic diversity of Trypanosoma evansi in buffalo based on internal transcribed spacer (ITS) regions. J. Vet. Med. Sci. 69(5), 487–493; doi:10.1292/jvms.69.487 Kumar, S., Stecher, G. and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger datasets. Mol. Biol. Evol. 33(7), 1870–1874; doi:10.1093/molbev/msw054 Mahmood, M.S. and Alobaidii, W.A. 2022. Molecular detection of Trypanosoma species in sheep and goats in Mosul city. Iraqi J. Vet. Sci. 36(2), 445–449; doi:10.33899/ijvs.2021.130488.1835 Mamman, S.A., Dakul, D.A., Yohanna, J.A., Dogo, G.A., Reuben, R.C., Ogunleye, O.O., Tyem, D.A., Peter, J.G. and Kamani, J. 2021. Parasitological, serological, and molecular survey of trypanosomosis (Surra) in camels slaughtered in northwestern Nigeria. Trop. Anim. Health Prod. 53(6), 537; https://orcid.org/0000-0002-7940-810X Musa, O., Zablon, K.N., Eucharia, U.K., Geoffrey, M.M., Ellie, O.O. and Daniel K M. 2005. Detection of trypanosomes in small ruminants and pigs in western Kenya: Important reservoirs in the epidemiology of sleeping sickness. Kinetoplastid Biol. Dis. 4(5), 1–7; doi: 10.1186/1475-9292-4-5 Mulenga, G.M., Namangala, B., Chilongo, K., Mubamba, C., Hayashida, K., Henning, L. and Gummow, B. 2021. Challenges in the Diagnostic Performance of Parasitological and Molecular Tests in the Surveillance of African Trypanosomiasis in Eastern Zambia. Trop. Med. Infect. Dis. 6(62), 68; doi:10.25903/1bw9-0s79 Rjeibi, M.R., Ben Hamida, T., Dalgatova, Z., Mahjoub, T., Rejeb, A., Dridi, W. and Gharbi, M. 2015. First report of surra (Trypanosoma evansi infection) in a Tunisian dog. Parasite 22(22), 3; doi:10.1051/parasite/2015004 Sadek, A., El-Khabaz, K., El-Genedy, S. and El-Gioushy, M. 2021. Comparative diagnostic performance of microscopic examination, polyclonal antigen-ELISA, and polymerase chain reaction for the detection of Trypanosoma evansi in Camels (Camelus dromedarius). Adv. Anim. Vet. Sci. 9, 1004; doi:10.17582/journal.aavs/202 Salim, B., Bakheit, M.A., Kamau, J., Nakamura, I. and Sugimoto, C. 2011. Molecular epidemiology of camel trypanosomiasis based on ITS1 rDNA and RoTat 1.2 VSG gene in the Sudan. Parasit Vectors 4, 31; doi:10.1186/1756-3305-4-31 SAS. 2012. Statistical analysis system, user’s guide. Statistical. Version 9.4. Cary, NC: SAS Institute Inc. Sivajothi, S., Rayulu, V.C., Malakondaiah, P. and Sreenivasulu, D. 2016. Diagnosis of Trypanosoma evansi in bovines by indirect ELISA. J. Parasitic Dis. 40(1), 141–144; doi:10.1007/s12639-014-0465-z Soulsby, E.J.L. 1982. Helminthes, arthropods and protozoa parasites of domesticated animals, London: Baillière Tindall, 232–233; doi:10.1016/0035-9203(84)90110-X Tariq, M., Badshah, F., Khan, M.S., Ibáñez-Arancibia, E., De Los Ríos-escalante, P.R., Khan, N.U., Naeem, S., Manzoor, A., Tahir, R., Mubashir, M., Ilyas, M., Manzoor, G.A. and Ben Said, M. 2024. Prevalence of trypanosomiasis caused by Trypanosoma evansi (Kinetoplastea, Trypanosomatidae) in domestic ruminants from Southern Punjab, Pakistan. Vet. World 17(9), 1955–1965; doi:10.14202/vetworld.2024.1955-1965 Taylor, K. and Authié, E.M.L. 2004. Pathogenesis of animal trypanosomiasis. In The Trypanosomiases. Eds., Holmes, P.H. and Miles, M.A. Wallingford, UK: CAB International, pp: 331–353; doi:10.1079/9780851994758.0331 Van den Bossche, P. and Delespaux, V. 2011. Options for the control of tsetse transmitted livestock trypanosomosis. An epidemiological perspective. Vet. Parasitol. 181, 37–42. | ||
| How to Cite this Article |
| Pubmed Style Faraj AA, Fadhil AI, Abed HH. Morpho-molecular characterization of Trypanosoma evansi in sheep. Open Vet. J.. 2025; 15(9): 4527-4532. doi:10.5455/OVJ.2025.v15.i9.58 Web Style Faraj AA, Fadhil AI, Abed HH. Morpho-molecular characterization of Trypanosoma evansi in sheep. https://www.openveterinaryjournal.com/?mno=251639 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i9.58 AMA (American Medical Association) Style Faraj AA, Fadhil AI, Abed HH. Morpho-molecular characterization of Trypanosoma evansi in sheep. Open Vet. J.. 2025; 15(9): 4527-4532. doi:10.5455/OVJ.2025.v15.i9.58 Vancouver/ICMJE Style Faraj AA, Fadhil AI, Abed HH. Morpho-molecular characterization of Trypanosoma evansi in sheep. Open Vet. J.. (2025), [cited January 25, 2026]; 15(9): 4527-4532. doi:10.5455/OVJ.2025.v15.i9.58 Harvard Style Faraj, A. A., Fadhil, . A. I. & Abed, . H. H. (2025) Morpho-molecular characterization of Trypanosoma evansi in sheep. Open Vet. J., 15 (9), 4527-4532. doi:10.5455/OVJ.2025.v15.i9.58 Turabian Style Faraj, Azhar Ali, Ali Issa Fadhil, and Howaida Hamel Abed. 2025. Morpho-molecular characterization of Trypanosoma evansi in sheep. Open Veterinary Journal, 15 (9), 4527-4532. doi:10.5455/OVJ.2025.v15.i9.58 Chicago Style Faraj, Azhar Ali, Ali Issa Fadhil, and Howaida Hamel Abed. "Morpho-molecular characterization of Trypanosoma evansi in sheep." Open Veterinary Journal 15 (2025), 4527-4532. doi:10.5455/OVJ.2025.v15.i9.58 MLA (The Modern Language Association) Style Faraj, Azhar Ali, Ali Issa Fadhil, and Howaida Hamel Abed. "Morpho-molecular characterization of Trypanosoma evansi in sheep." Open Veterinary Journal 15.9 (2025), 4527-4532. Print. doi:10.5455/OVJ.2025.v15.i9.58 APA (American Psychological Association) Style Faraj, A. A., Fadhil, . A. I. & Abed, . H. H. (2025) Morpho-molecular characterization of Trypanosoma evansi in sheep. Open Veterinary Journal, 15 (9), 4527-4532. doi:10.5455/OVJ.2025.v15.i9.58 |