| Research Article | ||
Open Vet. J.. 2025; 15(8): 3854-3861 Open Veterinary Journal, (2025), Vol. 15(8): 3854-3861 Research Article Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysisKatherine Bander Faraj1, Ali A. Al-Iedani1* and Waled Majeed Seger21Department of Microbiology, College of Veterinary Medicine, University of Basrah, Basrah, Iraq 2Department of Pathology and Poultry Diseases, College of Veterinary Medicine, University of Basrah, Basrah, Iraq *Corresponding Author: Ali A. Al-Iedani. Department of Microbiology, College of Veterinary Medicine, University of Basrah, Basrah, Iraq. Email: ali.eesa [at] uobasrah.edu.iq Submitted: 03/04/2025 Revised: 01/07/2025 Accepted: 12/07/2025 Published: 31/08/2025 © 2025 Open Veterinary Journal
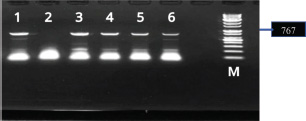
ABSTRACTBackground: Several Newcastle disease virus (NDV) genotypes are circulating worldwide, and the virus continually evolves, leading to more diversity and severity. Of several viral genotypes, VII is particularly important because it has been linked to recent ND outbreaks worldwide. Aim: This investigation aimed to diagnose the NDV from the broilers Ross 308 and to investigate the evolutionary relationship and molecular characteristics of the isolates that caused the outbreak during 2023–2024. Methods: Approximately 250 tissue samples were collected from broilers with different clinical signs and postmortem lesions related to NDV. The samples were diagnosed and analyzed using conventional polymerase chain reaction and sequencing analyses with bioinformatics. Results: Using polymerase chain reaction targeting the F gene, NDV was successfully detected in the examined samples. The NDV was isolated from geographical areas in four governorates. The global genetic diversity and epidemiological distribution of genotype VII of NDV in Iraq. Based on the 767-bp fusion gene sequences of the nine isolates of the present study and other reference viruses, further detailed characterization of the current Iraqi genotype VII and lentogenic or vaccine strains was performed. In addition, the phylogenetic study showed that the NDV strains of the current work (4FF, 7FF, 8FF, 10FF, 12FF, and 15FF) have an F protein cleavage site 112RRQKR↓ F117 and fit the new identified include sub-genotype VII 1.1. Conclusion: Our study explained that the virulent NDV strains are enzootic and are particularly widespread in Iraq. The ND isolates in this study also showed high similarity to Afghan and Iranian strains of genotype VII (97.95%–98.01% similarity). Keywords: Newcastle disease virus, Broilers, F gene, Pathogenicity, Phylogenetic tree. IntroductionNewcastle disease (ND) in chickens is caused by Newcastle disease virus (NDV), a paramyxovirus whose virulence varies considerably and results in different clinical manifestations. These infections range from highly fatal to subclinical or asymptomatic forms. For example, velogenic ND virus causes elevated mortality, in some cases up to 100%, although other strains, such as mesogenic or lentogenic viruses, can cause severe respiratory infections through opportunistic infection, or in environmental factors, we are faced with a negative situation (Alsahami et al., 2018). NDV has an extraordinary host variety, and there is evidence that each fowl species is at risk of contamination. The virus has been documented to infect at least 250 one-of-a-kind species of birds, making it one of the most host-adapted pathogens among avian viruses (Suarez et al., 2019). Genotypes VI and VII are further divided into seven (VIa-g) and five (VIIa-e) subgenotype, respectively. Genotypes V, VI, and VII of virulent viruses are the predominant genotypes circulating worldwide. Of these, genotype VII is particularly important given that it has been associated with many of the most recent outbreaks in Asia, Africa, and the Middle East (Dimitrov et al., 2019). The new classification system has grouped the entire VII (b+d+e+J+L) as one group assigned as VII.1.1 (Dimitrov et al., 2019). Though NDV is not highly pathogenic to humans, there is evidence that its potential impact is underestimated (Prajna et al., 2022). The hemagglutinin-neuraminidase and fusion (F) proteins are essential to the pathogenesis of NDV as they mediate access and exit of the virus into host cells. These proteins are key factors in the virus’s virulence, tissue tropism, and potential to reason disorder. The F protein is synthesized as an inactive precursor, F0, which should be cleaved into two subunits: F1, which includes the hydrophobic F peptide important for the F of virus and host membranes, and F2, which incorporates areas containing the Stabilize F complicated. This cleavage occurs with the aid of host mobile proteases and is critical for the activation of the F protein to mediate F (Zhang et al., 2023). A valuable molecular target for NDV genomes is the F gene, which plays an important role in viral infection and replication. It exhibits substantial variability for that of the F gene, providing significant information on epidemiological patterns and evolutionary relations among NDVs (da Silva et al., 2020). The amino acid sequence of the F protein cleavage site, comprising positions 112 to 117, is intimately linked to the virulence of NDV. This place is significant in controlling the virus potential to infect different tissues. The virulence of the NDV pressure is determined with the aid of the specific amino acids that dominate this location, including arginine or lysine, which are usually located at positions 112–116 in virulent traces, with phenylalanine at position 117 (112R/K-R-Q-K/R-R116↓F). Several basic residues render the F protein a target for common proteases determined in maximum tissues, including Furin. This tremendous protease penetration enables systemic virus replication and rapidly spreads the pathogen in the body. In contrast, single primary residues on the cleavage website are regularly found in less virulent strains, restricting proteolytic targeting and limiting infection (Panda et al., 2004; de Leeuw et al., 2005; Vahidi et al., 2023). In strains with low virulence, there are few basic residues that can often be separated by way of glutamic acid (E) or glycine (G). Leucine (L) is a residue at position 117. (112G/E-K/R-Q-G/E-R116↓L) is an example. Trypsin and different unique proteases restricted to positive tissues are required through those strains (e.g., the digestive or respiratory structures). These lines are typically used as vaccine traces and regularly motivate mild or subclinical disorders [e.g., A. LaSota and B1] (Panda et al., 2004; de Leeuw et al., 2005; Vahidi et al., 2023). Later, NDV became endemic in Iraq despite the intensive immunization program to control or prevent it, and it is still the main threat to the poultry industry in the country (Kadhim et al., 2024). The goal of this study was to apply reverse transcription polymerase chain reaction (RT-PCR) to diagnose and recognize NDV from infected broiler chickens in four Iraqi governorates. In addition to investigating the evolutionary relationship and molecular characteristics of the isolates that caused outbreaks during 2023–2024, intends to verify the genotypes of the isolates and analyze their phylogenetic relationships. Materials and MethodsSamplesThe tissue samples accrued under aseptic conditions from broiler farms(Ross 308) in Al-Basrah, Baghdad, Babil, Wasit, and Kirkuk governorates between July 2022 and March 2024 as shown in Figure 2. The clinical symptoms were observed as severe greenish diarrhea, depression, dehydration, loss of appetite, and facial swelling. About 250 samples were taken from 25 broiler farms in the middle and south of Iraq, each farm contains from (7,000 to 10,000) chicks. The high mortality rate started at 10% and reached 90% within a few days. A necropsy was performed on 75 samples of dead and dying birds. Suspect samples, including brain, liver, spleen, trachea, lung, proventriculus, and appendicular tonsils, were collected, and small pieces of tissue (2 g each) were cut with sterile forceps and scissors, assembled, and ground in an autoclaved pestle and mortar with the addition of liquid nitrogen before being stored at −20°C until used for virus isolation and identification. RNA extraction and cDNA synthesisViral RNA was obtained directly from harvested tissue using a total RNA isolation kit (Promega, USA) according to the manufacturer’s guidelines. The RNA concentration and purity of the samples were measured using a Nanodrop device, which determined the RNA concentration in ng/μl and the purity by measuring the optical density ratio. The reverse transcription system kit (catalog number A5000, Promega company, USA). Conventional polymerase chain reaction (PCR) for NDV detectionThe primer sequences are listed in Table 1 and were selected to amplify a 767-bp segment of the F gene containing the cleavage site. The partial sequence of such genes in the current study contained multiple basic amino acids, which are necessary for sequence with bioinformatics analyses, and also included some open reading frames. The reaction was exposed to 95°C for 5 minutes for initial denaturation, followed by 35 cycles of denaturation at 95°C for 20 seconds, annealing at 59.5°C for 20 seconds, and extension at 72°C for 1 minute with a final extension for 10 minutes at 68°C (Haryanto et al., 2015). The amplicons were loaded into 1% agarose gel stained with SYBR Safe DNA gel stain (Invitrogen, USA) for gel electrophoresis and visualized using a UV transilluminator (Aplegen, USA), Figure 1. Table 1. Sequence of Oligonucleotide Primers for Newcastle Disease Virus F Gene RT-PCR amplification.
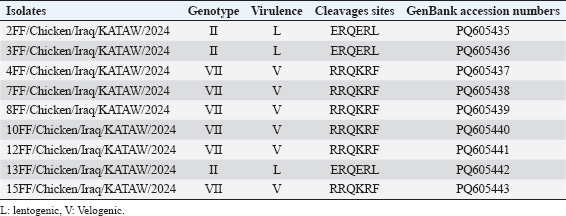
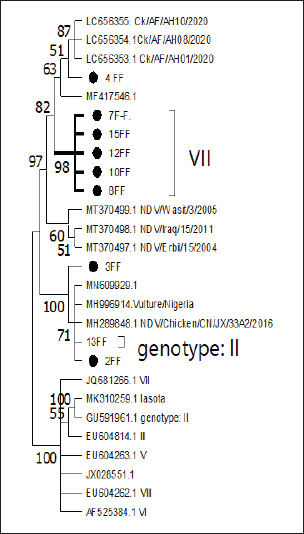
Nucleotide sequencing, amino acid analysis, and phylogenetic characterizationPCR products for the (25) representative governorates were removed from the gel of agarose gel and sent to Sanger sequencing (Korea). The strains that were identified by PCR and showed clear, clean, and single bands that represented all governorates were selected for sequence analyses. The same primers used for PCR amplification were also used for sequence analysis. Raw sequence data were edited and assembled manually using SeqMan Pro software (DNASTAR Lasergene, USA). Nucleotide and amino acid sequences were aligned to NDV strains representing each genotype retrieved from the GenBank database at NCBI. Sequence comparisons were performed using the BioEdit Sequence Alignment Editor (version 7.1.9) (Hall, 1999). Phylogenetic analysis was performed using MEGA version 6.06 using the neighbor-joining statistical method with the Kimura 2-parameter model and 1000 bootstrap replicates (Tamura et al., 2013). The phylogenetic tree was constructed using the F gene sequence data. Ethical approvalNot needed for this study. ResultsVirus detectionBetween 2023 and early 2024, two outbreaks of ND were recorded in broiler flocks in northern, central, and southern Iraq. The 150 positive samples in this study showed a clear band with specific primers, as shown in (Fig. 1). On the other hand, the DNA sequencing results revealed positive samples from different flocks. These strains were then subjected to genetic analysis. Phylogenetic analysis of NDVA partial analysis of the F gene sequences of the nine indicated Iraqi NDV field strains showed that all strains had varying degrees of identity with one another. Phylogenetic analysis of partial sequences of the designated strains demonstrated that the isolated viruses belong to (velogenic strain, genotype VII.1.1, and avirulent or vaccine strains). Only nine F gene sequences were included in the phylogenetic analysis and belonged to broilers in different governorates in Iraq (Baghdad, Basrah, Babil, Wasit, Kirkuk). Homology analysis of these viral genome sequences showed that the F genes had very high nucleotide sequence identity (98%–100%) with that from the gene bank. The isolates (4FF, 7FF, 8FF, 10FF, 12FF, and 15FF) belonged to genotype VII, while the nucleotide similarity (98%–99%) was consistent with that from Afghanistan (LC656353.1, LC656354, and LC656355). However, during the phylogenetic analysis of other representative isolates (2FF, 3FF, and 13FF) of avirulent or vaccine isolates, those belonging to genotype II had high F gene nucleotide similarity (98.83%–100%) with the strains isolated from Nigeria, Vietnam, and China (MH289851.1, MH996914.1, and MN609929) in 2003–2022. In addition, phylogenetic analysis based on the partial F gene sequence showed that the NDV strain (4FF, 7FF, 8FF, 10FF, 12FF, and 15FF) has an F protein cleavage site 112RRQKR↓ F117, which belongs to the newly identified genotype VII.1.1
Fig. 1. Agarose gel electrophoresis of RT-PCR products targeting the NDV fusion protein gene (767 bp); M marker,1,3,4,5 and 6 are positive samples 2; positive control, while 7 is negative. Genetic analysis of the F gene cleavage siteAmino acid analyses (proteolytic cleavage site) of the F gene revealed that the F0 precursor cleavage site of the nine Iraqi isolates from broilers contained multiple basic amino acid residues at the C-terminus of the F2 protein and a phenylalanine residue at the N-terminus of the F1 protein (112RRQKR|F117), which is a characteristic feature of velogenic strains of NDV (4FF, 7FF, 8FF, 10FF, 12FF, and 15FF). In comparison, the three isolates (2FF, 3FF, and 13FF) possessed the 112ERQER|L117 motif of the lentogenic strains of NDV, as shown in Table 2, Figures 3 and 4. Accession numbers of sequence dataThe nucleotide sequence data reported in this study were submitted to the GenBank sequence database and assigned accession numbers as in Table 2. DiscussionFrequent outbreaks of NDV present a critical risk to business fowl in Iraq, even within the lifestyles of ordinary vaccination packages (Khader et al., 2020). Secondary immunosuppressive infections that deteriorate the efficiency of vaccines are many of the environmental factors supposed to be answerable for the remaining power of NDV outbreaks in vaccinated populations (Bolha et al., 2013). Furthermore, the lack of herd immunity and subpar vaccination strategies should make the problem more severe, leading to inefficient disease management and the recurrent life of NDV (Yang et al., 1999). A polybasic amino acid F-protein cleavage site 112RRRKR↓F117, suggestive of a velogenic pathotype, was determined in six Iraq NDV isolates with Genbank accession numbers (PQ605437, PQ605438, PQ605439, PQ605440, PQ605441, and PQ605443), according to genetic evaluation of the partial sequences of the F protein gene. In addition, for the Iraqi NDV isolates, virulent cleavage motifs containing polybasic amino acids were registered by Al-Zuhariy et al. (2017) and Alazawy and Al Ajeeli (2020). Previously, ND was constrained to a few regions of Iraq. However, the disorder began in November 2010 and quickly spread throughout the country, killing a wide variety of commercial and rural chickens (Bakr, 2015). Instead of using the conventional tests of the mean death time test and intracerebral pathogenic index, several studies used F protein cleavage site sequence analyses to confirm the NDV pathotype (Gould et al., 2001) utilizing PCR products for epidemiological research and nucleotides of cleavage site analysis [Lin et al., (2003); Khan, et al., 2010].
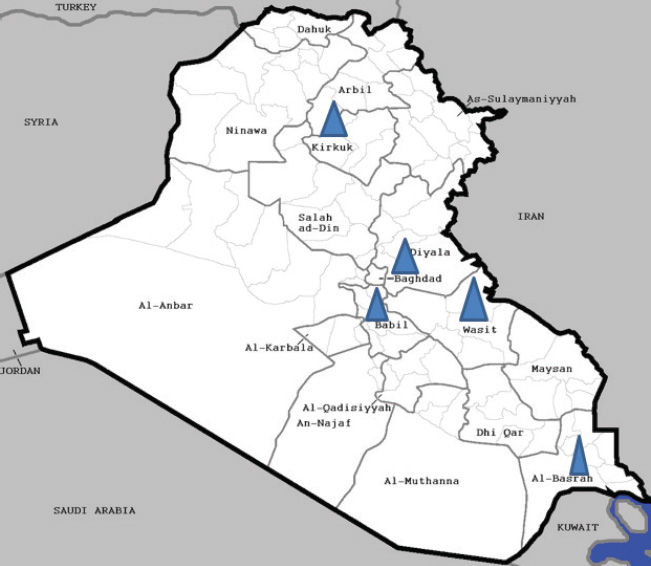
Fig. 2. Distribution of Iraqi NDV strains indicated in blue triangle in Basrah, Babil, Wasit, Kirkuk, Baghdad governorates. Table 2. NDV isolates, genotypes, and GenBank accession Numbers.
Fig. 3. Comparison of cleavage sites in NDV Isolates.
Fig. 4. Phylogenetic relationships of Iraqi NDV strains with reference strains from different countries based on the fusion gene. The strains indicated in black circles represent Study of Iraqi strains from middle and south of Iraq. Most field isolates clustered with sub-genotype VII1.1, distinct from commonly used vaccine strains such as LaSota.” Amino acid sequencing revealed the F1 protein; residue 117 showed phenylalanine (F) at the N-terminus, and the F2 protein showed 112RRQKRF117 at the C-terminus of all Iraqi virulent field NDV isolates. The amino acid sequence was similar to the motif installed in virulent NDV isolates (Khader et al., 2020). Since 1980, the epidemiological region of ND in Iraq has been characterized by periodic epizootics, whereas the summer in Iraq is usually characterized by enzootic ND infections. In (Fig. 4), the phylogenetic analysis of the currently selected Iraqi isolates in this study included the isolate can be divided into virulent groups that have a high similarity of 98% with (LC656353, LC656354, and LC656355) from Afghanistan and (MF417546) from Iran, causes of relatedness between our ND strains and that from gene bank or neighbor countries. These results are related to those of (Aliakbar Khabiri et al., 2023) who mentioned Phylogenetic and evolutionary distance analysis of F and hemagglutinin-neuraminidase genes clustered the virus in sub-genotype VII.1.1 in Iran, because some governorates of Iraq in this study, like Basrah, are located on the borderline of Iran. The rapidly spreading subgenotype VIId strains are considered major circulating ND viruses in many parts of the globe (Zhang et al., 2014). The phylogenetic tree (Fig. 4) also shows that the Iraqi isolates analyzed in this study are grouped within the virulent genotype VII. Among the reference sequences, MT370499, MT370498, and MT370497 are particularly significant. These sequences, which represent NDV strains from Iraq (NDV/Wasit/3/2005, NDV/Iraq/15/2011, and NDV/Erbil/15/2004), respectively, show and confirm high similarity to the Iraqi isolates. Their classification within these sequences is the virulent clade of genotype VII. The isolates (4FF, 7FF, 8FF, 10FF, 12FF, and 15FF) fall under clade VII because of their grouping in the tree. They have a close evolutionary relationship and are assigned to genotype VII. In addition, the tree showed the relatedness of the isolates to strains from different regions, such as NDV/Chicken/CN/JX/22F2/2017 (MH289851) from China, (MH996914. Vulture/Nigeria) and historical vaccine strains, such as Chicken/USA /LaSota /1946 (MH392212.1). The isolates (2FF, 3FF, and 13FF) are part of genotype II. The isolates (2FF and 13FF) are grouped within a subclade of genotype II, but 3FF appears to have some evolutionary distance from the others in genotype II, but still belongs to this genotype. This classification reflects their evolutionary divergence and genetic similarity, as shown in the tree. NDV genotype VII is the predominant genotype in poultry in the Middle East, with most NDV isolates from wild birds additionally assigned to this genotype (Kim et al., 2007). As routine vaccination programs against ND, including LaSota and B1 strains, have not been effective in Iraq, virulent NDV is still a big threat for Iraqi poultry industry. Previous studies have shown that compared with broilers vaccinated with heterologous vaccines LaSota, broilers vaccinated with homologous vaccines (against VII) had lower mortality and morbidity when challenged with virulent NDV (Hu et al., 2009). ConclusionThis study highlights the notable genetic diversity and geographic distribution of the NDV in the Iraqi broiler industry, with a focus on genotype VII, a prominent and virulent genotype associated with recent outbreaks that are globally significant. To verify the frequency of NDV in tissue samples collected from four Iraqi governorates, the researchers performed PCR and phylogenetic analyses. They identified a novel genotype VII.1.1 demonstrated that the strains they identified share a high level of genetic similarity with strains from Iran and Afghanistan, suggesting potential regional epidemiological links. This study highlights the enzootic presence of virulent NDV in Iraq, emphasizing the need for improved biosecurity measures, consistent surveillance, and tailored vaccination programs to lessen its spread and harmful effects on poultry health. AcknowledgmentI am deeply grateful to my supervisors for their encouragement throughout my research. In addition, I am thankful to the College of Veterinary Medicine, University of Basrah. Conflicts of interestRegarding the publishing of this article, the authors state that they have no conflicts of interest. FundingThe authors did not receive support from any organization for the submitted work (self-fund). Author contributionKatherine Bander Faraj: Writing original draft, Software, Resources, Methodology: samples collection and bioinformatics by Waleed Majeed. Supervision, All authors have read and agreed to the published version of the manuscript. Data availabilityAll data are available upon request. ReferencesAlazawy, A.K. and Al Ajeeli, K.S. 2020. Isolation and molecular identification of wild Newcastle disease virus isolated from broiler farms of Diyala Province, Iraq. Vet. World 13(1), 33–39. Alsahami, A., Ideris, A., Omar, A.R., Ramanoon, S.Z. and Sadiq, M.B. 2018. Seroprevalence of the Newcastle disease virus in backyard chickens and herd-level risk factors of Newcastle disease in poultry farms in Oman. Int. J. Vet. Sci. Med. 6(2), 186–191. Al-Zuhariy, M.T.B., Abdulmaged, S.H., Rabee, R.H. and Al-Baldawi, A.A.A. 2017. Isolation and identification of the Newcastle disease virus from field outbreaks in broiler and layer flocks in Iraq. Iraqi J. Vet. Med. 41(1), 23–27. Bakr, M.T. 2015. Effects of probiotics and immune modulators on the immune response Newcastle disease in the presence of E. coli as stress. PhD thesis, College of Veterinary Medicine, University of Baghdad, Iraq. Bolha, L., Benčina, D., Cizelj, I., Oven, I., Slavec, B., Rojs, O.Z. and Narat, M. 2013. Effects of coinfection of Mycoplasma synoviae and lentogenic Newcastle disease virus on cytokine and chemokine gene expression in chicken embryos. Poult. Sci. 92(12), 3134–3143. da Silva, A.P., Aston, E.J., Chiwanga, G.H., Birakos, A., Muhairwa, A.P., Kayang, B.B., Kelly, T., Zhou, H. and Gallardo, R.A. 2020. Molecular characterization of Newcastle disease virus genotype VII from Tanzania. Virol. J. 17, 30. De Leeuw, O.S., Koch, G., Hartog, L., Ravenshorst, N. and Peeters, B.P.H. 2005. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the hemagglutinin-neuraminidase protein. J. Gen. Virol. 86(Pt 6), 1759–1769. Dimitrov, K.M., Abolnik, C., Afonso, C.L., Albina, E., Bahl, J. and Berg, M. 2019. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 75, 103917. Gould, A.R., Kattenbelt, J.A., Selleck, P., Hansson, E., Della-Porta, A. and Westbury, H.A. 2001. Virulent Newcastle disease in Australia: a molecular epidemiological analysis of viruses isolated before and during the outbreaks of 1998–2000. Virus Res. 77(1), 51–60. Hakeem, J.K., Shlaga, A.K. and Ali, S.H. 2024. Clinical and molecular identification of Newcastle disease virus in naturally infected chicks in Thi-Qar province of Iraq. Open Vet. J. 14(11), 2817–2826. Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. Haryanto, A., Purwaningrum, M., Verawati, S., Irianingsih, S. and Wijayanti, N. 2015. Pathotyping of local isolates Newcastle disease virus from field specimens by RT-PCR and restriction endonuclease analysis. Procedia Chem. 14, 85–90. Hu, S., Ma, H., Wu, Y., Liu, W., Wang, X., Liu, Y. and Liu, X. 2009. A vaccine candidate for the attenuated Newcastle disease virus genotype VII generated by reverse genetics. Vaccine 27(6), 904–910. Kadhim, H.J., Shlaga, A.K. and Ali, S.H. 2024. Clinical and molecular identification of Newcastle disease virus in naturally infected chicks in Thi-Qar province of Iraq. Open Vet. J. 14(11), 2817–2826; doi: 10.5455/OVJ.2024.v14.i11.10 Khabiri, A., Toroghi, R., Mohammadabadi, M. and Tabatabaeizadeh, S.E. 2023. Introduction of a Newcastle disease virus challenge strain (sub-genotype VII.1.1) isolated in Iran. Vet. Res. Forum 14(4), 221–228; doi: 10.30466/vrf.2022.548152.3373 Khader, M., El-Kady, M. and Shaheed, I. 2020. Pathogenesis of Newcastle disease virus genotype VII in chickens vaccinated with LaSota and inactivated Newcastle disease vaccines. Int. J. Vet. Sci. 9(2), 105–110. Khan, T.A., Rue, C.A., Rehmani, S.F., Ahmed, A., Wasilenko, J.L., Miller, P.J. and Afonso, C.L. 2010. Phylogenetic and biological characterization of the Newcastle disease virus isolated from Pakistan. J. Clin. Microbiol. 48(5), 1892–1894. Kim, L.M., King, D.J., Curry, P.E., Suarez, D.L., Swayne, D.E., Stallknecht, D.E., Slemons, R.D., Pedersen, J.C., Senne, D.A., Winker, K. and Afonso, C.L. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81(22), 12641–12653. Lin, M.Y., Liu, H.J. and Ke, G.M. 2003. Genetic and antigenic analysis of Newcastle disease viruses from recent outbreaks in Taiwan. Avian Pathol. 32(4), 345–350. Panda, A., Huang, Z., Elankumaran, S., Rockemann, D.D. and Samal, K.S. 2004. Role of the fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 36(1), 1–10. Prajna, N.V., Lalitha, P., Chen, C., Zhong, L., Lietman, T.M., Doan, T. and Seitzman, G.D. 2022. Acute keratoconjunctivitis resulting from co-infection with avian Newcastle virus and human adenovirus. Cornea 41(5), 630–631. Salih, R.H., Odisho, M., Al-Shammari, A.M. and Ibrahim, O.M. 2017. Antiviral effects of Olea europaea leaves extract and interferon-beta on gene expression of Newcastle disease virus. Adv. Anim. Vet. Sci. 5(11), 436–445. Suarez, D.L., Miller, P.J., Koch, G., Mundt, E. and Rautenschlein, S. 2019. Newcastle disease, other avian paramyxoviruses, and avian metapneumovirus infections. In Diseases of poultry. Eds., Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., Wit, S., Grimes, T., Johnson, D., Kromm, M., Prajitno, T.Y., Rubinoff, I. and Zavala, G. Wiley, pp: 109–166. Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30(12), 2725–2729. Vahidi, V., Ebrahimi, S., Akbari, N. and Jafari, P. 2023. Molecular identification and phylogenetic study of a fusion gene of Newcastle disease virus isolated from broiler poultry farms in Markazi Province, Iran. Arch. Razi Inst. 78(6), 1794. Yang, C.Y., Shieh, H.K., Lin, Y.L. and Chang, P.C. 1999. Newcastle disease virus isolated from recent outbreaks in Taiwan phylogenetically related to viruses (genotype VII) from recent outbreaks in western Europe. Avian Dis. 43(1), 125–130; doi: 10.2307/1592467 Zhang, D., Ding, Z. and Xu, X. 2023. Pathologic mechanisms of the Newcastle disease virus. Viruses 15(4), 864. Zhang, Y., Shao, M., Yu, X., Zhao, J. and Zhang, G. 2014. Molecular characterization of chicken-derived genotype VIId Newcastle disease virus isolates in China during 2005–2012 reveals a new length of hemagglutinin-neuraminidase. Infect. Genet. Evol. 21, 359–366. | ||
| How to Cite this Article |
| Pubmed Style Faraj KB, Al-iedani AA, Seger WM. Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis. Open Vet. J.. 2025; 15(8): 3854-3861. doi:10.5455/OVJ.2025.v15.i8.51 Web Style Faraj KB, Al-iedani AA, Seger WM. Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis. https://www.openveterinaryjournal.com/?mno=250646 [Access: January 26, 2026]. doi:10.5455/OVJ.2025.v15.i8.51 AMA (American Medical Association) Style Faraj KB, Al-iedani AA, Seger WM. Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis. Open Vet. J.. 2025; 15(8): 3854-3861. doi:10.5455/OVJ.2025.v15.i8.51 Vancouver/ICMJE Style Faraj KB, Al-iedani AA, Seger WM. Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis. Open Vet. J.. (2025), [cited January 26, 2026]; 15(8): 3854-3861. doi:10.5455/OVJ.2025.v15.i8.51 Harvard Style Faraj, K. B., Al-iedani, . A. A. & Seger, . W. M. (2025) Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis. Open Vet. J., 15 (8), 3854-3861. doi:10.5455/OVJ.2025.v15.i8.51 Turabian Style Faraj, Katherine Bander, Ali A. Al-iedani, and Waled Majeed Seger. 2025. Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis. Open Veterinary Journal, 15 (8), 3854-3861. doi:10.5455/OVJ.2025.v15.i8.51 Chicago Style Faraj, Katherine Bander, Ali A. Al-iedani, and Waled Majeed Seger. "Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis." Open Veterinary Journal 15 (2025), 3854-3861. doi:10.5455/OVJ.2025.v15.i8.51 MLA (The Modern Language Association) Style Faraj, Katherine Bander, Ali A. Al-iedani, and Waled Majeed Seger. "Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis." Open Veterinary Journal 15.8 (2025), 3854-3861. Print. doi:10.5455/OVJ.2025.v15.i8.51 APA (American Psychological Association) Style Faraj, K. B., Al-iedani, . A. A. & Seger, . W. M. (2025) Genetic diversity and phylogenetic characterization of Newcastle Disease Virus in broilers in Iraq based on the F gene analysis. Open Veterinary Journal, 15 (8), 3854-3861. doi:10.5455/OVJ.2025.v15.i8.51 |