| Research Article | ||
Open Vet. J.. 2025; 15(9): 4482-4495
Open Veterinary Journal, (2025), Vol. 15(9): 4482-4495 Research Article Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomicsAnjar Windarsih1, Hendy Dwi Warmiko2, Yuny Erwanto3, Nor Kartini Abu Bakar4, Abdul Rohman5,6*, Ahmad Zainul Hasan2, Muhammad Khalid Abdullah2, Ade Marmita7, Hamidah Itha'atur Rif'ah7, Mustika Furi8 and Yuniar Khasanah11Research Center for Food Technology and Processing, National Research and Innovation Agency, Yogyakarta, Indonesia 2Corpora Science Research Laboratory, PT. Wiralab Analitika Solusindo, Yogyakarta, Indonesia 3Faculty of Animal Sciences, Universitas Gadjah Mada, Yogyakarta, Indonesia 4Department of Chemistry, Faculty of Science, Universiti Malaya, Kuala Lumpur, Malaysia 5Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Universitas Gadjah Mada, Yogyakarta, Indonesia 6Center of Excellence, Institute for Halal Industry and Systems, Universitas Gadjah Mada, Yogyakarta, Indonesia 7Halal Products Assurance Organizing Agency, Jakarta, Indonesia 8Sekolah Tinggi Ilmu Farmasi Riau, Kota Pekanbaru, Indonesia *Corresponding Author: Abdul Rohman. Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Universitas Gadjah Mada, Yogyakarta, Indonesia. Email: abdulkimfar [at] gmail.com Submitted: 27/02/2025 Revised: 25/07/2025 Accepted: 27/08/2025 Published: 30/09/2025 © 2025 Open Veterinary Journal
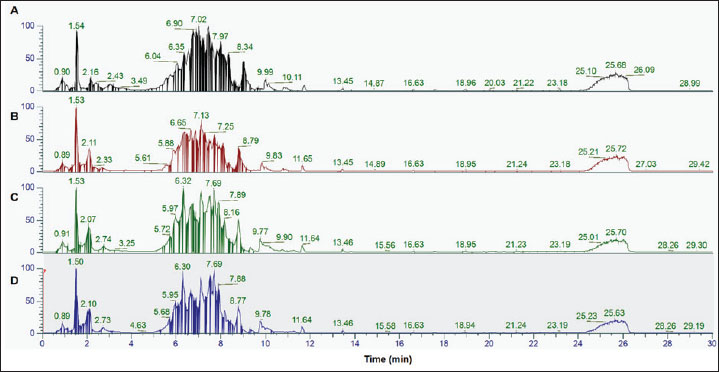
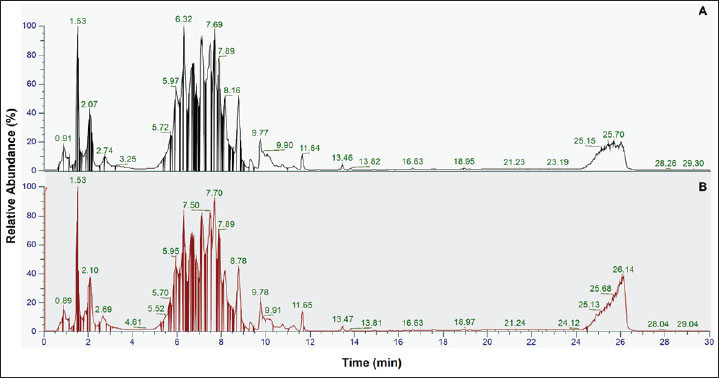
AbstractBackground: Detecting porcine gelatin in gelatin powders and food products is crucial for ensuring compliance with dietary restrictions, ethical considerations, and regulatory standards. Aim: This study presents a non-targeted proteomic approach utilizing liquid chromatography coupled with high-resolution Orbitrap mass spectrometry to identify and quantify porcine gelatin in gelatin powder and food products, such as gummy candies and marshmallows. Methods: Peptide sequences unique to bovine and porcine gelatin were identified through comparative mass spectrometry data analysis, enabling reliable differentiation from gelatin sourced from other animals. Both extraction buffers, including Tris-HCl buffer (pH 8.0) and ammonium bicarbonate (pH 8.0), were effective for protein and peptide extraction in gelatin. Several porcine gelatin peptide markers were obtained in both extraction buffers. In addition, this study revealed that the reduction and alkylation processes using dithiothreitol and iodoacetamide, respectively, are unnecessary, thereby simplifying the extraction steps. Results: This method was successfully tested with gelatin used in complex food matrices such as gummy candies and marshmallows. Porcine gelatin was detected in three samples of gummy candies and three samples of marshmallows. Conclusion: Therefore, future research, such as the targeted approach and standardization, is strongly required to obtain a valid, reliable, and reproducible method for the analysis of porcine gelatin in food and pharmaceutical product applications for halal authentication purposes. Keywords: Porcine gelatin, Bovine gelatin, Non-targeted proteomics, LC-HRMS, Halal authentication. IntroductionGelatin, a widely used biopolymer derived primarily from animal collagen, plays a critical role in the food, pharmaceutical, and cosmetic industries due to its gelling, thickening, and stabilizing properties. Porcine gelatin, which is sourced from pig-derived collagen, is one of the most commonly used types of gelatin due to its excellent functional properties and widespread availability (Zhang et al., 2024; Hassan et al., 2025a,b). However, the authenticity and quality of gelatin products, including those in gelatin powders and foods containing gelatin, are often subject to scrutiny due to concerns over adulteration, mislabeling, and contamination with other animal-derived ingredients (Uddin et al., 2021; Zhu et al., 2023). Therefore, the analysis of gelatin, particularly its source, composition, and authenticity, is essential for quality control and regulatory purposes across various industries, including food, pharmaceuticals, and cosmetics (Rohman et al., 2020). A DNA-based method, such as real-time polymerase chain reaction (RT-PCR), is commonly used to detect specific genetic markers or trace DNA from animal sources, such as porcine or bovine, within gelatin (Mohamad et al., 2018; Sultana et al., 2020). However, this technique has limitations in its applicability to processed gelatin. Since gelatin production involves collagen hydrolysis and denaturation, the DNA originally present in the collagen is typically destroyed during processing. This makes RT-PCR less effective for identifying gelatin sources in commercial products, especially when gelatin is present in processed food matrices (Yang et al., 2018; Han et al., 2022). Furthermore, RT-PCR cannot provide detailed information about gelatin peptide composition, which is crucial for identifying specific gelatin types or detecting adulteration (Yörük et al., 2024). The lack of specificity for processed gelatin forms a significant research gap in the accurate and reliable identification of gelatin types, particularly in complex food products (Hassan et al., 2025a,b). Therefore, the development of analytical techniques capable of high sensitivity and selectivity for detecting gelatin adulteration and gelatin-containing foods is highly necessary. Proteomics offers numerous advantages for the analysis of porcine gelatin (Suratno et al., 2023; Zhu et al., 2023). As porcine gelatin is derived from collagen found in pig connective tissues, it presents unique biochemical signatures that can be identified through proteomic methods, making it a powerful tool for both quality control and authenticity testing. Proteomics provides a detailed and comprehensive profile of the peptide and protein composition of gelatin (Hassan et al., 2025a,b; Kwon et al., 2025). Using proteomics, researchers can identify these peptides and map them to specific regions of the collagen molecule, allowing for the precise characterization of the gelatin source. This is particularly important for distinguishing porcine gelatin from other animal-derived gelatins, such as bovine or porcine gelatin (Cai et al., 2021; Sidira et al., 2024). Proteomics can identify unique peptide markers specific to porcine gelatin, providing a reliable method for determining its authenticity in raw or processed gelatin products (Garcia-Vaquero and Mirzapour-Kouhdasht, 2023). Unlike traditional methods, such as PCR, which are specific to certain genetic markers, proteomics provides a more flexible and comprehensive approach, capable of identifying a wide range of peptides without requiring predetermined markers (Kwon et al., 2025). This allows researchers to uncover novel peptide biomarkers that may not have been previously identified, offering new insights into the properties and potential adulterants of gelatin. Proteomics, particularly when coupled with liquid chromatography-high-resolution mass spectrometry (LC-HRMS), offers numerous advantages for the analysis of porcine gelatin (Varunjikar et al., 2024). LC-HRMS is a powerful analytical technique that provides high sensitivity and specificity, making it particularly effective in the proteomics analysis of gelatin (Aini et al., 2023). In the context of gelatin analysis, LC-HRMS allows for the precise identification and quantification of peptides derived from gelatin, even at low concentrations (Harlina et al., 2024). The combination of liquid chromatography (LC) with high-resolution mass spectrometry (MS) enables the separation of complex peptide mixtures, followed by accurate mass determination, which is crucial for distinguishing between peptides with similar molecular weights (Aydoğan, 2020). The high resolution of LC-HRMS enhances its ability to identify peptide markers specific to gelatin, which are critical for verifying the gelatin’s origin (such as bovine gelatin), assessing its purity, and detecting any contaminants or adulterants (Zhu et al., 2023). Additionally, the technique offers excellent specificity, ensuring that the analysis can pinpoint unique peptide sequences associated with gelatin’s hydrolyzed products. This level of detail is essential for proteomic studies that aim to profile the protein structure and integrity of gelatin in various applications, including food safety, pharmaceutical formulations, and quality control processes (Carrera et al., 2024). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) has emerged as a powerful tool for protein analysis, offering high sensitivity and specificity. Previous studies have successfully employed LC-MS/MS to identify and quantify porcine gelatin in gelatin powder and foods containing gelatin (Zhu et al., 2023). For instance, LC-MS/MS with various digestion methods has been developed for the authentication of confectionary products by identifying porcine gelatin markers (Dewi et al., 2023). This approach demonstrated high accuracy in differentiating porcine gelatin from bovine sources. Similarly, LC-MS/MS was successfully used in conjunction with chemometric analysis to classify porcine and bovine gelatin based on their unique peptide profiles (Salamah et al., 2019). However, all previously reported methods require rigorous sample preparation, including complex preparation steps with long LC-MS/MS measurement running times. Building upon these advancements, this study aims to develop a rapid and efficient method for the analysis of porcine gelatin in gelatin powder and foods containing gelatin using sophisticated liquid chromatography coupled with high-resolution Orbitrap mass spectrometry (LC-Orbitrap HRMS). Our study aimed to develop a more efficient method capable of effectively detecting peptide markers of porcine gelatin. First, we developed a simple sample preparation technique to simplify the protein extraction steps by removing several steps, thereby reducing the chemical reagents used and shortening the extraction time. Moreover, we developed a short running time using LC-HRMS Orbitrap, which has superior sensitivity and selectivity and is capable of detecting very low levels of porcine gelatin in gummy candy and marshmallow samples. Materials and MethodsChemicalsLC-MS grade acetonitrile, LC-MS grade water, flexi for positive and negative calibration solutions, and proteomic grade trypsin were obtained from Fisher Scientific (Thermo Scientific, Rockford, USA). Tris-base, iodoacetamide (IAA), dithiothreitol (DTT), and ammonium bicarbonate were purchased from Sigma-Aldrich (USA). HCl and NaOH were purchased from Merck (Darmstadt, Germany) for analysis. Sample collectionThe Gelatin standard, consisting of bovine gelatin and porcine gelatin, was obtained from Sigma-Aldrich (USA). Five gummy candies and four marshmallow samples from different brands were purchased from several markets in Jakarta and Yogyakarta, Indonesia. The commercial samples were randomly selected. Protein extractionProtein extraction was performed by referring to the method outlined by Zhu et al. (2023) with modifications. For the gelatin standard, 500 mg of gelatin standard was weighed and placed into a 50 ml centrifuge tube. The sample was added to 25 ml of extraction buffer and vortexed for 10 seconds. In this study, two types of buffers were used: Tris-HCl buffer pH 8.0 ± 0.2 and ammonium bicarbonate pH 8.0 ± 0.2. The mixtures were then sonicated at 50oC for 1 hour. After the sonication process was completed, the samples were centrifuged at 7,000 g for 6 minutes at room temperature (25oC). Subsequently, 500 µl of the supernatant was pipetted into a 1.5 ml centrifuge tube. The supernatant was added with 500 µl of cold acetone (−20oC). For gummy candy and marshmallow samples, 2 g samples were weighed and placed into 15 ml centrifuge tubes. The samples were added with Tris-HCl buffer pH 8.0 ± 0.2 for protein extraction and vortexed for 10 seconds. Following the vortex, the samples were processed using the same steps as those for the gelatin standard. After the addition of 500 µl of cold acetone (−20oC), the samples were kept at −20oC for 30 minutes to complete protein precipitation. Protein digestionThe digestion process was performed according to the method described by Windarsih et al. (2022) with slight modifications. A total of 50 µl of the reconstituted supernatant from the gelatin standard and samples was collected and placed into a 1.5 ml centrifuge tube. The solution was added with 50 µl digestion buffer of ammonium bicarbonate, pH 8.0 ± 0.2, and vortexed for 10 seconds. After this step, 4 μl of 50 mM DTT was added for reduction at 75°C for 30 minutes at 75oC. Then, 9 μl of 50 mM IAA was added for alkylation and incubated for 30 minutes in the dark. We also performed the digestion process without the addition of DTT and IAA for the porcine gelatin standard. Subsequently, 5 µl of proteomic-grade trypsin (20 µg/100 µg) dissolved in 1 mM HCl was added to the solution for protein digestion. The mixture was then vortexed for 10 seconds. The mixture was then incubated using a thermomixer at 37oC for 16 hours. Following the incubation process, the reaction was stopped using 10 µl formic acid and vortexed for 10 seconds. Water was added until the volume reached 1 ml. The mixture was then filtered using a 0.22 µm nylon filter placed into a 2-ml HPLC vial for LC-HRMS analysis. LC-HRMS untargeted proteomics analysisThe digested proteins were analyzed using ultra-high performance LC (Thermo Scientific Vanquish, Germany) with a high-resolution mass spectrometer detector (Orbitrap Exploris 240, Thermo Scientific, Bremen Germany). The peptides in both the gelatin standard and samples were separated using Acclaim PepMap C18 (150 × 2.1 mm × μm) using a gradient elution technique. 5 μl of the sample was injected and eluted using water (A) and acetonitrile (B) mobile phases. The temperature of the sampler was set at 8oC. To increase the resolution, 0.1% of both mobile phases was added and sonicated for 30 minutes for degassing purposes. The duration of peptide elution using LC was 30 minutes, as follows: 4% B (0–2.0 minutes), 15% B (2.1–15.0 minutes), 60% B (15.1–20.0 minutes), 60% B (20.1–22.0 minutes), 90% B (22.1–24.1 minutes), 4% B (24.1–24.2 minutes), and 4% B (24.2–30.0 minutes). Peptides were eluted and separated within the column, which was maintained at 40oC. The separated peptides were passed to HRMS through the interface. The ESI was operated in positive ionization mode. The scanning process to detect the peptides was conducted in the scan range of 300–2,000 m/z with a full scan resolution of 90,000 FWHM. Moreover, fragmentation (MS2) was performed at a resolution of 22,500. During the process, the temperature of the capillary was set at 300oC. Before performing the HRMS analysis, the mass spectrometer was calibrated using both positive and negative calibration solutions to ensure the validity and accuracy of the mass (m/z) measurement. Data analysisThe total ion chromatogram (TIC) obtained by LC-HRMS was exported using Xcalibur software (Thermo Scientific, USA). The TIC was further analyzed using Proteome Discoverer software (Thermo Scientific, USA) to identify the proteins and peptides contained in the gelatin standards and samples. Both porcine and bovine FASTA proteins were downloaded from uniport.org and imported into Proteome Discoverer. The proteins and peptides contained in the samples were analyzed against FASTA. Then, the peptide markers of bovine and porcine gelatin were identified based on previous publications. Peptide markers were selected on the basis of high-confidence MS/MS spectra, strict mass accuracy thresholds (<5 ppm), and consistent retention times. Any initially ambiguous features were excluded unless they showed reproducible, species-specific patterns across replicates. Ethical approvalNot needed for this study. ResultsProteomics analysis in the gelatin standardUntargeted proteomics using LC-Orbitrap HRMS was successfully used to identify proteins and peptides in bovine and porcine gelatin standard gelatin. The method could effectively and efficiently detect and analyze the proteins and peptides from the gelatin standard through only 30 minutes of LC separation. Figure 1 shows the TIC of bovine and porcine gelatin standards extracted using two different buffers of Tris-HCl (pH 8.0) and ammonium bicarbonate (pH 8.0).
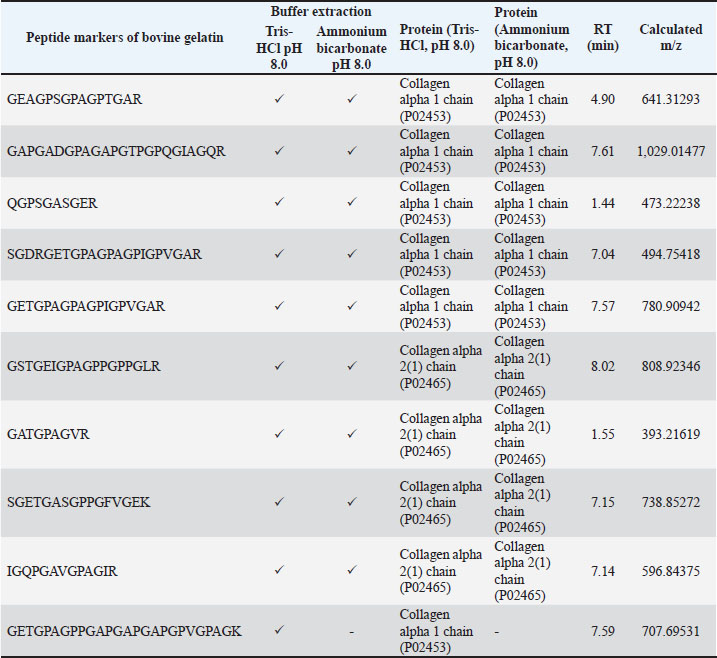
Fig. 1. The TIC of the gelatin standard was extracted using different extraction buffers: bovine gelatin-Tris HCl buffer pH 8.0 (A), bovine gelatin-ammonium bicarbonate pH 8.0 (B), porcine gelatin-Tris HCl buffer pH 8.0 (C), and porcine gelatin-ammonium bicarbonate pH 8.0 (D). Identification of peptide markers using bovine gelatin standardsThe identification of peptide markers from bovine gelatin standards is a critical process in the quality control and characterization of gelatin-derived products. To identify proteins and peptides in gelatin, LC-HRMS with Orbitrap mass analyzer was used. Table 1 shows the peptide markers found in bovine gelatin obtained from two different extraction buffers. In general, Tris-HCl (pH 8.0) and ammonium bicarbonate (pH 8.0) had similar performance in extracting peptides from bovine gelatin. Ten peptide markers of bovine gelatin were obtained using Tris-HCl (pH 8.0), whereas nine peptide markers were found in bovine gelatin using ammonium bicarbonate (pH 8.0). The peptides were obtained from the collagen alpha 1 chain and collagen alpha 2 chain. For all peptides, the same source of protein was found using Tris-HCl (pH 8.0) and ammonium bicarbonate (pH 8.0). Therefore, both extraction solvents resulted in similar results of peptide markers from bovine gelatin, except for the peptide GETGPAGPPGAPGAPGAPGPVGPAGK, which was only detected using Tris-HCl (pH 8.0). Table 1. Identification of peptide markers from bovine gelatin standard obtained from two different buffer extractions of Tris-HCl (pH 8.0) and ammonium bicarbonate (pH 8.0).
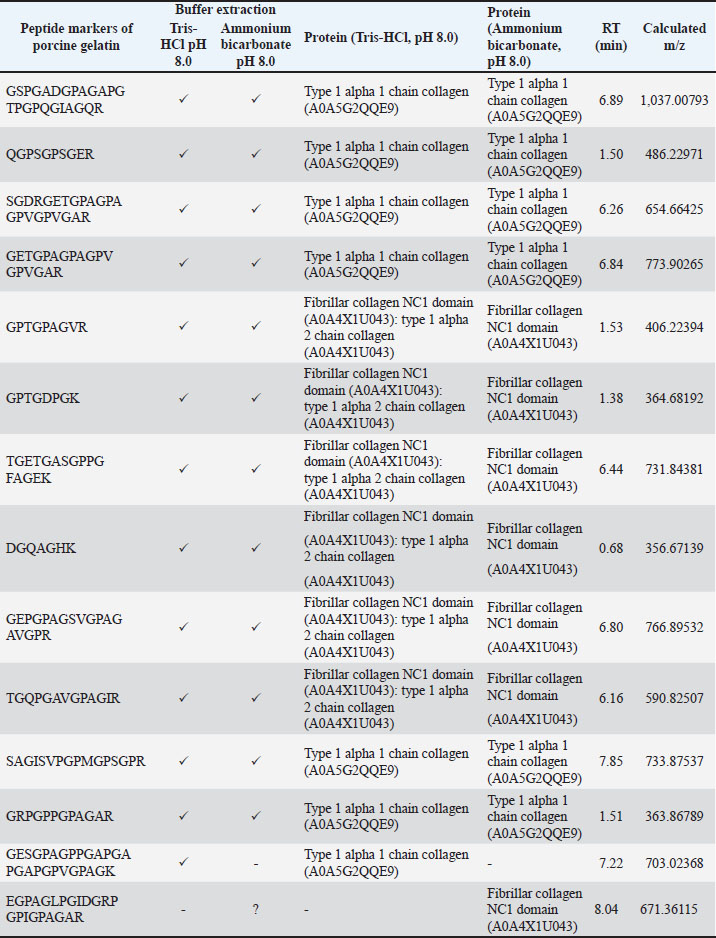
Identification of peptide markers using porcine gelatin standardsThe obtained peptide markers of porcine gelatin are shown in Table 2. The results showed that both Tris-HCl buffer (pH 8.0) and ammonium bicarbonate (pH 8.0) successfully detected porcine gelatin peptide markers with comparable performance. Proteins were extracted from the collagen alpha 1 chain and fibrillar collagen. The same number of 13 porcine gelatin peptide markers could be detected using both extraction solvents. However, there was a difference in the peptide of GESGPAGPPGAPGAPGAPGPVGPAGK, which was only detected in Tris-HCl buffer (pH 8.0). This peptide belongs to the Sus scrofa collagen type 1 alpha 1 chain. In contrast, the peptide marker of EGPAGLPGIDGRPGPIGPAGAR, which belongs to the fibrillar collagen protein NC1 domain, was only detected in porcine gelatin extracted using ammonium bicarbonate (pH 8.0). Table 2. Identification of peptide markers from porcine gelatin standard obtained from two different buffer extractions of Tris-HCl (pH 8.0) and ammonium bicarbonate (pH 8.0).
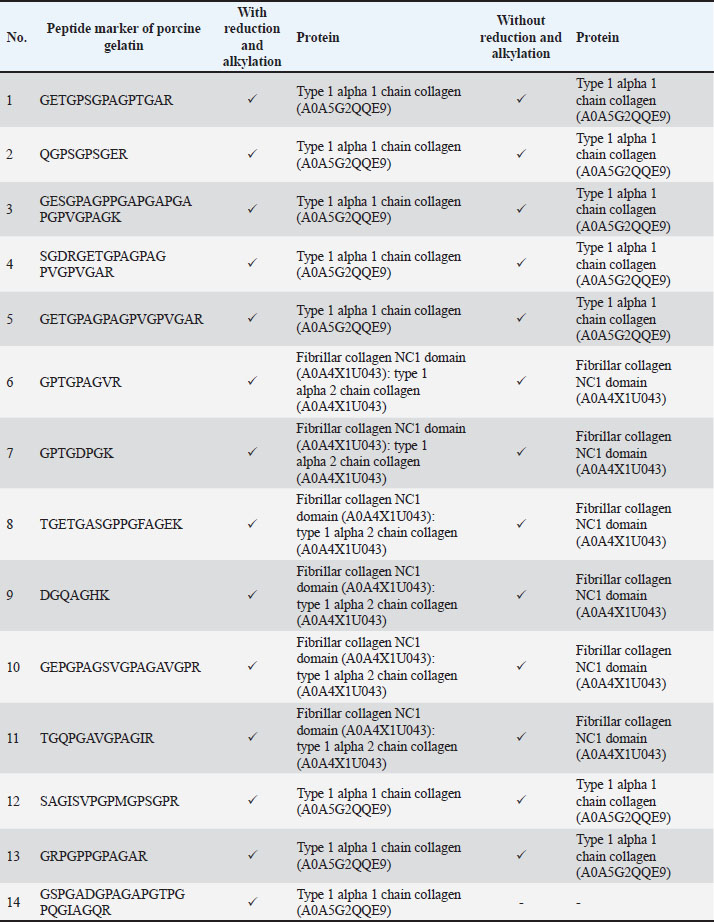
Protein digestionIn this study, porcine gelatin was subjected to simplified techniques by reducing several steps in protein digestion. The reduction step using DTT and the alkylation step using IAA were removed in the process to investigate the effectiveness of protein digestion without reduction and alkylation steps. According to the TIC (Fig. 2), the sample of porcine gelatin without reduction and alkylation steps demonstrated a similar TIC pattern to those applied reduction and alkylation steps. Table 3 shows the comparison of peptide markers in porcine gelatin extracted using Tris-HCl buffer (pH 8.0) with and without the use of DTT and IAA for reduction and alkylation, respectively. The removal of DTT and IAA in the extraction steps did not significantly affect the peptide markers detected in the porcine gelatin samples. Porcine gelatin samples extracted without DTT and IAA demonstrated the same peptide markers as those extracted using DTT and IAA. These results suggest/indicate that DTT and IAA are not necessary for protein extraction in gelatin. Table 3. Peptide markers of porcine gelatin standard obtained from two different digestion processes: reduction using DTT and alkylation using IAA process and without reduction and alkylation process.
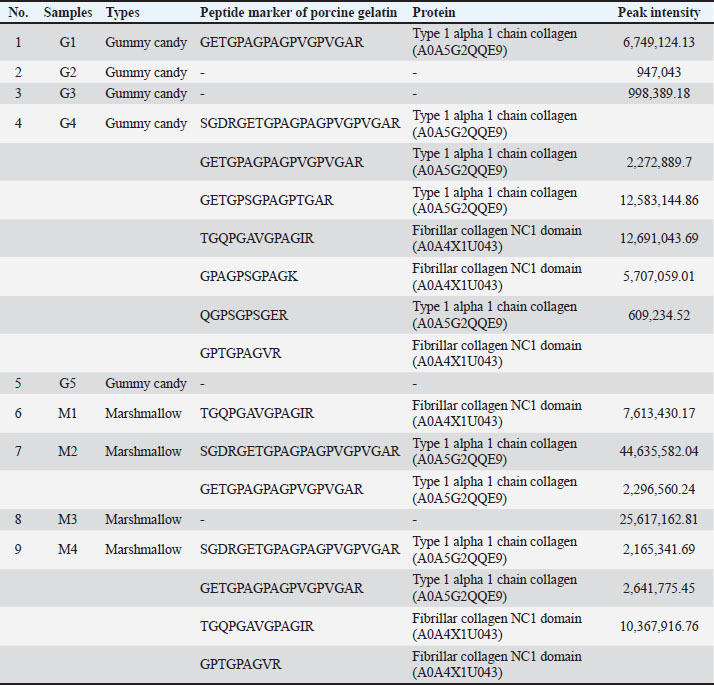
Fig. 2. TIC of porcine gelatin standard obtained from the process with reduction using DTT and alkylation using IAA steps (A) and without reduction and alkylation steps (B). Gelatin analysis of gummy candy and marshmallow samplesIn this study, LC-HRMS detected porcine gelatin in several samples of marshmallow and gummy candies obtained from markets. Table 4 shows the results of the analysis of commercial gummy candy and marshmallow samples obtained from different brands. Among the five gummy candies samples tested, two were positive for porcine gelatin. In addition, two marshmallow samples were found to contain porcine gelatin. Table 4. Identification of porcine gelatin peptide markers in commercial gummy candies and marshmallow samples.
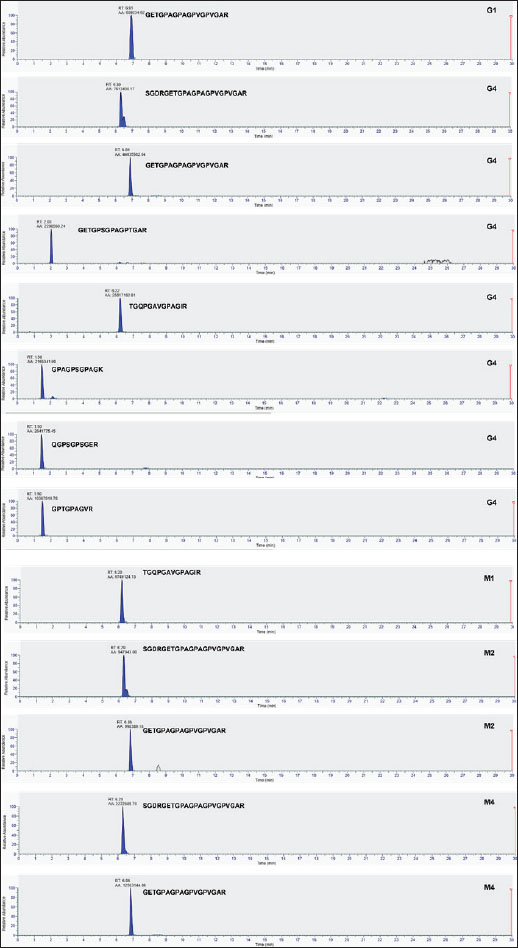
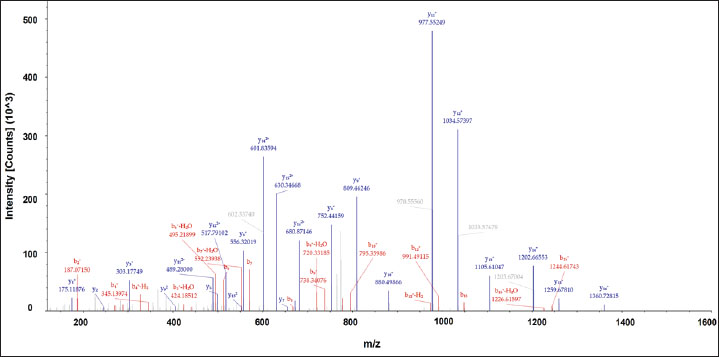
DiscussionThe detection of porcine gelatin is quite challenging due to some processes involved in gelatin production. The identification of collagens and their peptides would be very useful for investigating the gelatin source required for authentication. Figure 1 shows the TIC of bovine and porcine gelatin standards extracted using two different buffers of Tris-HCl (pH 8.0) and ammonium bicarbonate (pH 8.0). The method could effectively and efficiently detect and analyze the proteins and peptides from gelatin standard only through 30 minutes of LC separation. In this study, after the TICs were processed using the Proteome Discoverer software to reveal the proteins and peptides obtained from LC-HRMS analysis, the peptide markers were identified from the gelatin by referring to previously published papers (Zhang et al., 2009; Zhu et al., 2023). Two buffers were used to obtain the most effective buffer for gelatin protein extraction. Tris-HCl (pH 8.0) and ammonium bicarbonate (pH 8.0) are the most commonly used buffers for protein extraction from gelatin. The TIC showed similar patterns between the porcine and bovine samples, indicating the high similarity of the protein contents in the bovine and porcine gelatin. It might happen because collagen, such as collagen alpha 1 and collagen alpha 2 chains, is the major protein contained in both bovine and porcine gelatin. Identification of peptide markers using bovine gelatin standardsThe identification of peptide markers from bovine gelatin standards is a critical process in the quality control and characterization of gelatin-derived products. Bovine gelatin, a protein derived from bovine collagen, is commonly used in the food, pharmaceutical, and cosmetic industries. Characterization of peptide markers involves analyzing hydrolyzed peptides that result from gelatin breakdown, often through enzymatic digestion or chemical processes (Hamid et al., 2020). Techniques such as MS, LC-MS/MS, and proteomics-based approaches are often employed to identify these peptide markers. Researchers can detect any adulteration, ensure product authenticity, and assess the functional properties of gelatin by analyzing the molecular weight, amino acid composition, and fragmentation patterns of these peptides. In this study, LC-HRMS with Orbitrap mass analyzer was used to identify proteins and peptides in gelatin. LC-HRMS allows for the precise identification and quantification of peptides derived from gelatin, even at low concentrations, due to its superior sensitivity and specificity (Ovbude et al., 2024). Table 1 shows the peptide markers found in bovine gelatin obtained from two different extraction buffers. In general, Tris-Hcl (pH 8.0) and ammonium bicarbonate (pH 8.0) had similar performance in extracting peptides from bovine gelatin. Ten peptide markers of bovine gelatin were obtained using Tris-HCl (pH 8.0), whereas nine peptide markers were found in bovine gelatin using ammonium bicarbonate (pH 8.0). The peptides were obtained from the collagen alpha 1(1) chain (P02453) and collagen alpha 2(1) chain (P02465). Both extraction buffers contained the same protein source for each detected peptide. Therefore, both extraction solvents resulted in similar peptide marker results from bovine gelatin. Identification of peptide markers using porcine gelatin standardsThe identification of peptide markers from porcine gelatin standards is a crucial step in ensuring the authenticity, traceability, and quality of gelatin used in food, pharmaceuticals, and cosmetics (Abdullah Sani et al., 2021). The use of advanced proteomic techniques, particularly LC-MS/MS, is one of the key aspects in identifying peptide markers from porcine gelatin. This method allows the separation and identification of peptides based on their mass-to-charge ratio, enabling the detection of distinct peptide markers that are unique to porcine gelatin. Various porcine-specific peptides have been identified as reliable markers for the analysis of gelatin (Zhang et al., 2009; Zhu et al., 2023). The obtained peptide markers of porcine gelatin are shown in Table 2. The results showed that both Tris-HCl buffer (pH 8.0) and ammonium bicarbonate (pH 8.0) successfully detected porcine gelatin peptide markers with comparable performance. The peptides of GPTGPAGVR, GPTGDPGK, TGETGASGPPGFAGEK, DGQAGHK, GEPGPAGSVGPAGAVGPR, and TGQPGAVGPAGIR were detected from the protein sources of fibrillar collagen NC1 domain (A0A4X1U043) and collagen type 1 alpha 2 chain (A0A4X1U043) using Tris-HCl pH 8.0. In contrast, ammonium bicarbonate (pH 8.0) could only detect these peptides from the fibrillar collagen NC1 domain (A0A4X1U043). The contrasting results highlight the critical role of buffer composition in protein handling and downstream analysis, especially for complex and structured proteins, such as collagen. The Tris-HCl pH 8.0 buffer likely provided a more favorable environment for the collagen type I alpha 2 chain, either by enhancing its solubility, stabilizing its structure, or optimizing the efficiency of peptide generation/detection, allowing for the successful identification of the peptides from this source (Zhu et al., 2023). Gelatin analysis using simplified methodsIn this study, porcine gelatin was subjected to simplified techniques by reducing several steps in protein digestion. The reduction step using DTT and the alkylation step using IAA were removed in the process to investigate the effectiveness of protein digestion without reduction and alkylation steps. According to the TIC (Fig. 2), the sample of porcine gelatin without reduction and alkylation steps demonstrated a similar TIC pattern to those applied reduction and alkylation steps. Table 3 shows the comparison of peptide markers in porcine gelatin extracted using Tris-HCl buffer (pH 8.0) with and without DTT and IAA. The removal of DTT and IAA in the extraction steps did not significantly affect the peptide markers detected in the porcine gelatin samples. Porcine gelatin samples extracted without DTT and IAA demonstrated the same peptide markers as those extracted using DTT and IAA. These results suggest that DTT and IAA are not necessary for protein extraction in gelatin. Similarly, a previous study reported that reduction using DTT and alkylation using IAA are not necessary in the analysis of collagen because the disulfide bonds contained in collagen are only present at the end of the polypeptide chain, specifically within the propeptide. These disulfide bonds were hydrolyzed during the processing of gelatin (Yang et al., 2018). Moreover, other research also reported that the need for reduction and alkylation, typically used to break and modify disulfide bonds, is not a requirement for gelatin because disulfide bonds are largely removed or significantly reduced during the production of gelatin. The gelatin process primarily involves the breakdown of the triple-helix structure of collagen into smaller peptides, which does not necessitate further modifications to disulfide linkages (Müller and Winter, 2017). Gelatin analysis of gummy candy and marshmallow samplesGelatin plays a pivotal role in the food industry, particularly in the production of gummy candies and marshmallows. It is the key ingredient responsible for these products’ texture, structure, and elasticity. As such, the accurate identification and quantification of gelatin in gummy candies and marshmallows are crucial for quality control, regulatory compliance, and product authenticity. Both gummy candies and marshmallows are complex food matrices composed of gelatin, sugar, glucose syrup, starch, and other stabilizing agents. These ingredients can interfere with gelatin extraction and analysis, making it challenging to isolate and identify gelatin peptides. In gummy candies, gelatin is often present in relatively high concentrations; however, its interactions with other components, such as sugar and starch, can create a heterogeneous environment that complicates its detection. Similarly, in marshmallows, the gelatin may undergo partial denaturation due to the high temperature and whipping process, which can affect its molecular integrity and impact the reliability of analysis methods. LC-HRMS is a powerful tool for identifying gelatin peptides with high sensitivity and specificity, even in complex food samples. LC-MS/MS can effectively separate and analyze gelatin peptides from other food components, such as sugars, starches, and proteins, by exploiting differences in their mass-to-charge ratios in gummy candy and marshmallow samples. In this study, LC-HRMS detected porcine gelatin in several samples of marshmallow and gummy candies obtained from markets using Tris-HCl pH 8.0 for protein extraction. Table 4 shows the results of the analysis of commercial gummy candy and marshmallow samples obtained from different brands. Figure 3 shows the peak of each peptide marker found in Table 4. Among the five gummy candies samples tested, two were positive for porcine gelatin. The peptide GETGPAGPAGPVGPVGAR (collagen type 1 alpha 1 chain; accession number A0A5G2QQE9) was detected in both gummy candy samples. Among the four commercial marshmallow samples, three samples contained porcine gelatin. Among the two positive marshmallow samples, the peptide GETGPAGPAGPVGPVGAR was also detected. The peptide sequence GETGPAGPAGPVGPVGAR has been identified as a stable peptide marker in gelatin samples. Making it a reliable marker for identifying gelatin sources. This stability is essential for ensuring the accuracy of gelatin source identification in various products (Kwon et al., 2025). Figure 4 illustrates the fragmentation spectra of the GETGPAGPAGPVGPVGAR peptide. The results of this research indicated that the developed method could detect peptide markers of porcine gelatin from both types of samples, indicating the superior performance of LC-HRMS in detecting porcine gelatin in complex food samples. In addition, the developed extraction method could extract porcine gelatin from food samples containing complex matrices.
Fig. 3. Peak intensity of porcine peptide marker in gummy candies (G1 and G4) and marshmallow (M1, M2, and M4) samples.
Fig. 4. Fragmentation spectra (MS2 spectra) of GETGPAGPAGPVGPVGAR, a peptide marker of porcine gelatin. Currently, the DNA-based method using RT-PCR is still the standard method for detecting the authenticity of gelatin (Gina et al., 2024). However, some studies have reported that RT-PCR often provides false results for detecting porcine gelatin. Using the DNA-based method, the extraction step is crucial because the presence of DNA in gelatin is very low due to the high processing steps applied to produce gelatin (Jannat et al., 2018; Yörük et al., 2024). Therefore, it might cause biased information related to the source of gelatin, resulting in invalid information regarding the halal authenticity status of gelatin and foods containing gelatin. Therefore, our developed method using LC-HRMS-based proteomics is highly promising as a powerful technique for the analysis of porcine gelatin and gelatin-containing foods. Limitations and prospectsDespite the advancements in LC-MS/MS for gelatin analysis, several challenges remain. The presence of sugar, fat, and stabilizers can inhibit the extraction of gelatin peptides from gummy candies and marshmallows. The degradation of gelatin during high-temperature processing (such as in marshmallow production) can also affect the quality of peptide markers, resulting in incomplete or inconsistent identification. Additionally, the complexity of the peptide profiles in processed food samples may require advanced data analysis methods and robust databases to accurately assign peptide sequences to gelatin components. Future research should focus on optimizing extraction protocols to improve peptide recovery and minimize interference from other ingredients. Furthermore, efforts to develop comprehensive peptide databases for different gelatin sources and processing conditions could enhance proteomic analysis accuracy and reliability. The integration of LC-MS/MS with other analytical techniques, such as infrared spectroscopy or nuclear magnetic resonance, could provide complementary data and improve the overall understanding of gelatin behavior in food systems. ConclusionUntargeted proteomics using LC-Orbitrap HRMS provides sensitive detection for the analysis of peptide markers of porcine and bovine gelatin in gelatin powder and foods containing gelatin. In this study, a simpler sample preparation technique was successfully developed to extract protein and peptides by reducing the number of steps and the use of chemical reagents. Moreover, a short gradient in LC was efficiently developed to separate peptides in gelatin, thereby providing an effective method in terms of analysis time with optimum results. Two types of extraction solvents, including Tris-HCl buffer (pH 8.0) and ammonium bicarbonate (pH 8.0), had good capability to extract proteins and peptides from gelatin with comparable results. This developed method was effectively applied to detect porcine gelatin in commercial gummy candies and marshmallow samples. This study proposed a powerful method for the analysis of porcine gelatin in gelatin powder and foods containing gelatin with simplified extraction techniques and short analysis time using LC-orbitrap high-resolution MS. This method is a promising standard method for the analysis of porcine gelatin for quality control and halal authentication of gelatin powder and foods containing gelatin. Therefore, further research using a broader range of food products is important. In addition, a targeted proteomics approach (quantitative proteomics analysis), validation methods, and standardization are required to obtain a valid, reliable, and reproducible method for the analysis of porcine gelatin in food and pharmaceutical products. AcknowledgmentThe authors acknowledge Corpora Science Research Laboratory for facilitating the LC-HRMS Orbitrap Exploris 240 instrument. The authors would like to thank Badan Penyelenggara Jaminan Produk Halal (BPJPH), Laboratorium BRIN Kawasan Yogyakarta, and Laboratorium Penelitian dan Pengujian Terpadu (LPPT) UGM for supporting this research. Conflict of interestThe authors declare no conflicts of interest associated with this research. FundingThis research was supported by the RIIM LPDP Grant and BRIN, grant number 172/IV/KS/11/2023 and 6815/UN1/DITLIT/Dit-Lit/KP.01.03/202. Author contributionsAW, HDW, YE, YK, NKAB, AR: conceptualization and design, drafted the manuscript MKA, AZH, AM: formal analysis and interpretation data, revised and edited the manuscript HIR: acquisition and participated in the preparation and critical checking of the manuscript. MF: edited the references and checking the manuscript for grammatical errors. All authors have read and agreed to the publication of the final version of the manuscript. Data availabilityAll data were provided in the manuscript. ReferencesAbdullah Sani, M.S., Ismail, A.M., Azid, A. and Samsudin, M.S. 2021. Forensic food models for the authentication and quantification of porcine adulterant in gelatin and marshmallow. Food Control 130, 108350; doi:10.1016/j.foodcontrol.2021.108350 Aini, S.R., Rohman, A., Erwanto, M., Handayani, A. and Irnawati. 2023. Metabolomics approach used for Halal authentication analysis of food and pharmaceutical products: a review. Food Res. 7(3), 180–187. Aydoğan, C. 2020. Recent advances and applications in LC-HRMS for food and plant natural products: a critical review. Anal. BioAnal. Chem. 412(9), 1973–1991; doi:10.1007/S00216-019-02328-6 Cai, S., Jiang, M., Zhao, K., Huang, X., Fei, F., Cao, B., Cui, X., Duan, J., Zhao, M., Han, S. and Liu, R. 2021. Quantitative strategy of ultrasound-assisted digestion combined with ultraperformance liquid chromatography-mass spectrometry (UPLC-MS/MS) for rapid identification of species-specific peptide markers in the application of food gelatin authentication. LWT - Food Sci. Technol. 147, 111590; doi:10.1016/J.LWT.2021.111590 Carrera, M., Abril, A.G., Pazos, M., Calo-Mata, P., Villa, T.G. and Barros-Velázquez, J. 2024. Proteins and peptides: proteomics approaches for food authentication and allergen profiling. Curr. Opinion Food Sci. 57, 101172; doi:10.1016/J.COFS.2024.101172 Dewi, K.R., Kusnandar, F., Yuliana, N.D., Ismayati, M., Solihat, N.N., Riantana, H. and Heryani, H. 2023. Application of LC-MS/MS coupled with various digestion methods for the identification of porcine gelatin markers in confectionery matrices. Indonesian J. Halal Res. 5(2), 53–66; doi:10.15575/IJHAR.V5I2.21191 Garcia-Vaquero, M. and Mirzapour-Kouhdasht, A. 2023. Proteomic and genomic biomarkers for gelatin source authentication: challenges and future outlook. Heliyon 9(6), e16621; doi:10.1016/J.HELIYON.2023.E16621 Gina, S., Rahmagiarti, C., Ummah, I.M., Sumantri, C., Suparto, I.H. and Darmawan, N. 2024. Assessment of commercial DNA extraction kits for porcine gelatin detection using RT-PCR and ddPCR. Sci. Technol. Indonesia 9(3), 605–612; doi:10.26554/STI.2024.9.3.605-612 Hamid, A.H., Fadzillah, N.A., Sani, M.S.A., Muhammad, N.W.F., Othman, R. and Rohman, A. 2020. Discrimination of porcine and bovine gelatines based on reducing sugar types in the Maillard reaction. Food Res. 4(2), 301–306; doi:10.26656/fr.2017.4(2).297 Han, S., Yan, Z., Huang, X., Cai, S., Zhao, M., Zheng, Y., Liu, X., Xu, H., Xie, Y., Hou, R., Duan, J.A. and Liu, R. 2022. Response boosting-based approach for absolute quantification of gelatin peptides using LC-MS/MS. Food Chem. 390, 133111; doi:10.1016/j.foodchem.2022.133111 Harlina, P.W., Maritha, V., Shahzad, R., Rafi, M., Geng, F., Musfiroh, I., Muchtaridi, M., Wahab, R., Al-Khedhairy, A.A., Koerniati, S. and Amalina, N.N. 2024. Comprehensive profiling and authentication of porcine, bovine, and goat bone gelatins through UHPLC-HRMS metabolomics and chemometric strategies. LWT - Food Sci. Technol. 205, 116529; doi:10.1016/J.LWT.2024.116529 Hassan, M., Kanwal, T., Siddiqui, A.J., Ali, A., Hussain, D. and Musharraf, S.G. 2025b. Authentication of porcine, bovine, and fish gelatins based on quantitative amino acid profile and chemometric analysis. Food Control 168, 110909; doi:10.1016/J.FOODCONT.2024.110909 Hassan, H.M., Souka, U.D. and Hassan, S.M. 2025a. Differentiation and quantification of bovine and pork gelatin using ultra-performance liquid chromatography-quantitative time-of-flight and ATR-FTIR spectroscopy: addressing challenges in mixed gelatin analysis and detection. Food Chem. 464, 141883; doi:10.1016/j.foodchem.2024.141883 Jannat, B., Ghorbani, K., Shafieyan, H., Kouchaki, S., Behfar, A., Sadeghi, N., Beyramysoltan, S., Rabbani, F., Dashtifard, S. and Sadeghi, M. 2018. Gelatin speciation using real-time PCR and mass spectrometry-based proteomics dataset analysis. Food Control 87, 79–87; doi:10.1016/J.FOODCONT.2017.12.006 Kwon, J., Shin, D., Park, G.W., Lee, G., Lee, E. and Kang, H.S. 2025. Comprehensive quantitative LC-MS/MS method for rapid gelatin source identification in food products: comparison with polymerase chain reaction. Food Res. Int. 201, 115611; doi:10.1016/J.FOODRES.2024.115611 Mohamad, N.A., Mustafa, S., Mokhtar, N.F. and El Sheikha, A.F. 2018. Molecular beacon-based real-time PCR for detecting porcine DNA in gelatin and gelatin capsules. J. Sci. Food Agric. 98(12), 4570–4577; doi:10.1002/JSFA.8985 Müller, T. and Winter, D. 2017. Systematic evaluation of protein reduction and alkylation reveals massive unspecific side effects of iodine-containing reagents. Mol. Cell. Proteomics MCP. 16(7), 1173; doi:10.1074/MCP.M116.064048 Ovbude, S.T., Sharmeen, S., Kyei, I., Olupathage, H., Jones, J., Bell, R.J., Powers, R. and Hage, D.S. 2024. Application of chromatographic methods in metabolomics: a review. J. Chromatog. B 1239, 124124; doi:10.1016/J.JCHROMB.2024.124124 Rohman, A., Windarsih, A., Erwanto, Y. and Zakaria, Z. 2020. Analytical methods for the analysis of porcine gelatin in food and pharmaceutical products for Halal authentication. Trends Food Sci. Technol. 101, 122–132; doi:10.1016/J.TIFS.2020.05.008 Salamah, N., Erwanto, Y., Martono, S., Maulana, I. and Rohman, A. 2019. Differentiation of bovine and porcine gelatines by lc-ms/ms and chemometrics. Int. J. Appl. Pharm. 11(4), 159–163; doi:10.22159/ijap.2019v11i4.30248 Sidira, M., Smaoui, S. and Varzakas, T. 2024. Recent proteomics, metabolomics, and lipidomics approaches for meat safety, processing, and quality analysis. Appl. Sci. 14(12), 5147; doi:10.3390/APP14125147 Sultana, S., Hossain, M.A.M., Azlan, A., Johan, M.R., Chowdhury, Z.Z. and Ali, M.E. 2020. TaqMan probe-based multiplex quantitative PCR assay for determination of bovine, porcine and fish DNA in gelatin admixture, food products and dietary supplements. J. Biotechnol. Chem. Biotechnol. Phys. Biotechnol. Food Chem. 325, 126756; doi:10.1016/j.foodchem.2020.126756 Suratno, Windarsih, A., Warmiko, H. D., Khasanah, Y., Indrianingsih, A.W. and Rohman, A. 2023. Metabolomics and proteomics using LC-Orbitrap HRMS for the detection of Pork in Tuna meat for Halal authentication. Food Anal. Methods 16, 867–877. doi: 10.1007/s12161-023-02472-x Uddin, S.M.K., Hossain, M.A.M., Sagadevan, S., Al Amin, M. and Johan, M.R. 2021. Halal and Kosher gelatin: applications and detection approaches with challenges and prospects. Food Biosci. 44, 101422; doi:10.1016/J.FBIO.2021.101422 Varunjikar, M.S., Lie, K.K., Lundebye, A.K., Belghit, I., Ørnsrud, R., Berntssen, M.G.H., Lecrenier, M.C., Oveland, E., Palmblad, N.M. and Rasinger, J.D. 2024. Proteomics for food and feed authentication in the circular food chain. Trends Food Sci. Technol. 153, 104710; doi:10.1016/J.TIFS.2024.104710 Windarsih, A., Suratno, Warmiko, H.D., Indrianingsih, A.W., Rohman, A. and Ulumuddin, Y.I. 2022. Untargeted metabolomics and proteomics approach using liquid chromatography-Orbitrap high-resolution mass spectrometry to detect pork adulteration in meat from Pangasius hypothalamus. Food Chem. 386, 132856; doi:10.1016/j.foodchem.2022.132856 Yang, C.T., Ghosh, D. and Beaudry, F. 2018. Detection of gelatin adulteration using bio-informatics, proteomics, and high-resolution mass spectrometry. Food Addit. Contaminants-Part. A. Chem. Anal. Control. Exposure Risk Assessment 35(4), 599–608; doi:10.1080/19440049.2017.1416680 Yörük, N.G., Yılmaz, F. and Soycan, A. 2024. Verification analyses for the detection of bovine and porcine species in foods containing animal gelatin with the Q-exactive ORBITRAP device. Food Anal. Methods 1, 1–12; doi:10.1007/S12161-024-02717-3/METRICS Zhang, W., Li, M., Chen, J., Chen, Y., Liu, C. and Wu, X. 2024. Modified gelatin: physicochemical properties, modification methods, and applications in food. J. Agricult. Food Chem. 72(38), 20705–20721. Zhang, G., Liu, T., Wang, Q., Chen, L., Lei, J., Luo, G., Ma, G. and Su, Z. 2009. Mass spectrometric detection of marker peptides in gelatin tryptic digests: a new method to differentiate between bovine and porcine gelatin. Food Hydrocoll. 23(7), 2001–2007; doi:10.1016/J.FOODHYD.2009.03.010 Zhu, X., Gu, S., Guo, D., Huang, X., Chen, N., Niu, B. and Deng, X. 2023. Determination of porcine-derived components in gelatin and gelatin-containing foods by high-performance liquid chromatography-tandem mass spectrometry. Food Hydrocoll. 134, 107978; doi:10.1016/J.FOODHYD.2022.107978 | ||
| How to Cite this Article |
| Pubmed Style Windarsih A, Warmiko HD, Erwanto Y, Bakar NKA, Rohman A, Hasan AZ, Abdullah MK, Marmita A, Rifah HI, Furi M, Khasanah Y. Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics. Open Vet. J.. 2025; 15(9): 4482-4495. doi:10.5455/OVJ.2025.v15.i9.54 Web Style Windarsih A, Warmiko HD, Erwanto Y, Bakar NKA, Rohman A, Hasan AZ, Abdullah MK, Marmita A, Rifah HI, Furi M, Khasanah Y. Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics. https://www.openveterinaryjournal.com/?mno=244913 [Access: January 24, 2026]. doi:10.5455/OVJ.2025.v15.i9.54 AMA (American Medical Association) Style Windarsih A, Warmiko HD, Erwanto Y, Bakar NKA, Rohman A, Hasan AZ, Abdullah MK, Marmita A, Rifah HI, Furi M, Khasanah Y. Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics. Open Vet. J.. 2025; 15(9): 4482-4495. doi:10.5455/OVJ.2025.v15.i9.54 Vancouver/ICMJE Style Windarsih A, Warmiko HD, Erwanto Y, Bakar NKA, Rohman A, Hasan AZ, Abdullah MK, Marmita A, Rifah HI, Furi M, Khasanah Y. Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics. Open Vet. J.. (2025), [cited January 24, 2026]; 15(9): 4482-4495. doi:10.5455/OVJ.2025.v15.i9.54 Harvard Style Windarsih, A., Warmiko, . H. D., Erwanto, . Y., Bakar, . N. K. A., Rohman, . A., Hasan, . A. Z., Abdullah, . M. K., Marmita, . A., Rifah, . H. I., Furi, . M. & Khasanah, . Y. (2025) Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics. Open Vet. J., 15 (9), 4482-4495. doi:10.5455/OVJ.2025.v15.i9.54 Turabian Style Windarsih, Anjar, Hendy Dwi Warmiko, Yuny Erwanto, Nor Kartini Abu Bakar, Abdul Rohman, Ahmad Zainul Hasan, Muhammad Khalid Abdullah, Ade Marmita, Hamidah Ithaatur Rifah, Mustika Furi, and Yuniar Khasanah. 2025. Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics. Open Veterinary Journal, 15 (9), 4482-4495. doi:10.5455/OVJ.2025.v15.i9.54 Chicago Style Windarsih, Anjar, Hendy Dwi Warmiko, Yuny Erwanto, Nor Kartini Abu Bakar, Abdul Rohman, Ahmad Zainul Hasan, Muhammad Khalid Abdullah, Ade Marmita, Hamidah Ithaatur Rifah, Mustika Furi, and Yuniar Khasanah. "Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics." Open Veterinary Journal 15 (2025), 4482-4495. doi:10.5455/OVJ.2025.v15.i9.54 MLA (The Modern Language Association) Style Windarsih, Anjar, Hendy Dwi Warmiko, Yuny Erwanto, Nor Kartini Abu Bakar, Abdul Rohman, Ahmad Zainul Hasan, Muhammad Khalid Abdullah, Ade Marmita, Hamidah Ithaatur Rifah, Mustika Furi, and Yuniar Khasanah. "Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics." Open Veterinary Journal 15.9 (2025), 4482-4495. Print. doi:10.5455/OVJ.2025.v15.i9.54 APA (American Psychological Association) Style Windarsih, A., Warmiko, . H. D., Erwanto, . Y., Bakar, . N. K. A., Rohman, . A., Hasan, . A. Z., Abdullah, . M. K., Marmita, . A., Rifah, . H. I., Furi, . M. & Khasanah, . Y. (2025) Analysis of porcine gelatin in gelatin powder and gelatin-containing foods using non-targeted LC-Orbitrap HRMS-based proteomics. Open Veterinary Journal, 15 (9), 4482-4495. doi:10.5455/OVJ.2025.v15.i9.54 |