| Research Article | ||
Open Vet. J.. 2025; 15(5): 2193-2205 Open Veterinary Journal, (2025), Vol. 15(5): 2193-2205 Original Research Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in SurabayaAzhari Athaillah Sulaiman1, Mustofa Helmi Effendi2, Leonardo Reza Srimaryanto1, Aswin Rafif Khairullah3, Pitaloka Kurniasari1, Wiwiek Tyasningsih4*, Ikechukwu Benjamin Moses5, Riza Zainuddin Ahmad3, Saifur Rehman6, Budiastuti Budiastuti7, Daniah Ashri Afnani8, Sheila Marty Yanestria9, Katty Hendriana Priscilia Riwu10 and Agus Widodo111Master Program in Veterinary Science and Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 2Division of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 3Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia 4Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 5Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, Nigeria 6Department of Pathobiology, Faculty of Veterinary and Animal Sciences, Gomal University, Dera Ismail Khan, Pakistan 7Study Program of Pharmacy Science, Faculty of Health Science, Universitas Muhammadiyah Surabaya, Surabaya, Indonesia 8Department of Microbiology and Parasitology, Faculty of Veterinary Medicine, Universitas Pendidikan Mandalika, Mataram, Indonesia 9Faculty of Veterinary Medicine, Universitas Wijaya Kusuma Surabaya, Surabaya, Indonesia 10Department of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Pendidikan Mandalika, Mataram, Indonesia 11Department of Health, Faculty of Vocational Studies, Universitas Airlangga, Surabaya, Indonesia *Corresponding Author: Wiwiek Tyasningsih. Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia. Email: wiwiek-t [at] fkh.unair.ac.id Submitted: 23/02/2025 Revised: 15/04/2025 Accepted: 22/04/2025 Published: 31/05/2025 © 2025 Open Veterinary Journal
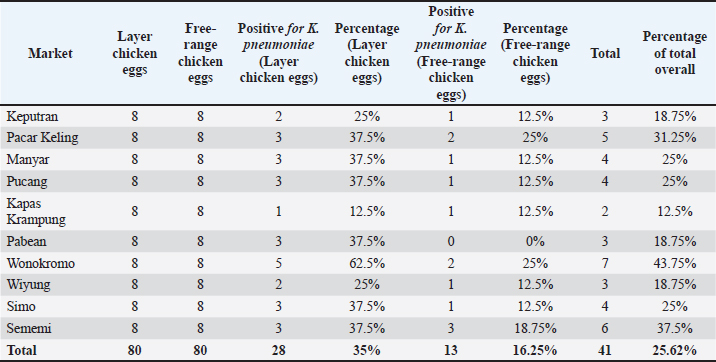
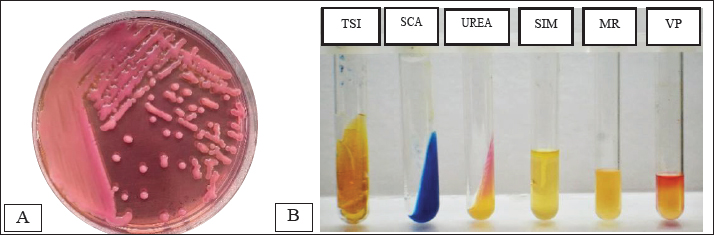
ABSTRACTBackground: Contamination with Klebsiella pneumoniae in food ingredients, including eggs, causes various dangers because it threatens public health, because it acts as a multidrug resistance (MDR) bacteria, especially the extended-spectrum beta-lactamase (ESBL) strain. The ESBL blaSHV gene is part of a broad-spectrum ESBL that is often found in Gram-negative bacteria. Aim: This study aimed to identify the ESBL blaSHV gene in K. pneumoniae MDR from chicken eggshells. Methods: This study used 160 samples of chicken eggshell swabs isolated on 1% BPW media from 10 traditional Surabaya markets. Samples were isolated using MCA media and were identified using Gram staining and biochemical tests. Detection of MDR using Muller-Hinton Agar. Results: Confirmation of ESBL in multidrug-resistant (MDR) isolates was performed using polymerase chain reaction to detect ESBL genes. The results showed that the isolation and identification of K. pneumoniae bacteria were 25.62% (41/160). Amoxicillin antibiotics showed the highest level of resistance at a percentage of 100% (41/41), followed by antibiotic resistance to erythromycin (90.24% (37/41), Streptomycin antibiotics were 26.82% (11/41), ciprofloxacin (14.63% (6/41), and Tetracycline antibiotic resistance was 7.31% (3/41). The results of MDR from K. pneumoniae showed 34.14% (14/41) of the isolates were then tested by PCR, which showed positive results for the blaSHV gene of 71.42% (10/14). Conclusion: The data from this study confirm the existence of K. pneumoniae bacteria isolated from egg shell swabs carrying the blaSHV gene from MDR isolates. Keywords: K. pneumoniae, MDR, ESBL, public health, egg shell. IntroductionA major global public health concern is food safety because dangerous microbes in food can cause a wide range of illnesses (Widodo et al., 2023; Kendek et al., 2024). Eggs are one of the food components that are commonly contaminated, even though eggs are a significant source of nutrition that delivers high-quality protein, necessary amino acids, vitamins, and minerals, and may be obtained in traditional markets and supermarkets (Kuang et al., 2018). Klebsiella pneumoniae is one of the bacteria that can infect eggs (Kahin et al., 2024). According to research by El Ftouhy et al. (2022), contamination of chicken egg shells by Klebsiella spp. in traditional markets and supermarkets was 24% and 4%, respectively. The contamination of chicken eggs by K. pneumoniae in traditional markets may result from both vertical and horizontal transmission (El Ftouhy et al., 2022). Additional research has demonstrated that agricultural surroundings, farm worker swabs, and egg swab samples are contaminated with K. pneumoniae bacteria (Dela et al., 2023). Gram-negative K. pneumoniae bacteria, which are members of the Enterobacteriaceae family, are frequently found on water surfaces, garbage, plants, soil, and other environmental surfaces. They can produce infectious agents in both humans and animals (Dameanti et al., 2023). Klebsiella pneumoniae is one of the bacteria that is resistant to a number of medications worldwide (Riwu et al., 2022). The prevalence of K. pneumoniae-related resistance has increased to 70% worldwide, and the mortality rate from infections has also increased to 40%–70% (Bradley and Scoular, 2021). According to recent studies, K. pneumoniae isolates have been identified as extensively drug-resistant (XDR) and produce extended-spectrum β-lactamase (ESBLs) and carbapenemases (Owaid and Al-Ouqaili, 2025). Although rarely recognized as a cause of foodborne illness, the presence of resistant and potentially virulent K. pneumoniae in food can pose health risks. This bacterium can be transmitted directly or spread resistance genes to other bacteria in food or the human gut (Hartantyo et al., 2020). One of the most urgent concerns for global public health in the twenty-first century is antimicrobial resistance (AMR) (Ferdinand et al., 2024). AMR is the result of an evolutionary process in which bacteria, fungi, viruses, and parasites develop resistance to antimicrobial medications, such as antibiotics, which are frequently used to treat infections (Salam et al., 2023). Contamination of food products with resistant bacteria occurs because animals and plants that serve as food sources for humans can act as vectors for the transfer of antimicrobial-resistant bacteria and AMR genes to humans. In addition, direct contact between humans and infected animals or environments, or with biological substances they produce, such as blood, urine, feces, saliva, and semen, can contribute to an increase in AMR (Chang et al., 2015). One of the various ways that bacteria develop resistance to antibiotics is by transferring resistance genes through plasmids (Benz et al., 2021; Khairullah et al., 2024). The genes blaSHV, blaTEM, and blaCTX-M are found on the chromosome or plasmid and regulate the β-lactamase enzyme manufacturing (Effendi et al., 2018; Pourhossein et al., 2020). The ESBL enzyme, which is commonly found in K. pneumoniae, is responsible for 20% of plasmid-mediated penicillin group resistance, specifically SHV (Yuwono, 2013). The Gram-negative bacteria Enterobacteriaceae produce extended-spectrum beta-lactamases, which are enzymes that hydrolyze penicillins as well as first, second, and third-generation cephalosporins and the aztreonam group (Ansharieta et al., 2021). These enzymes are inhibited by beta-lactamase inhibitors such as clavulanate, sulbactam, and tazobactam (Ghafourian et al., 2015). Hussein et al. (2024) concluded that the high levels of antibiotic resistance observed in K. pneumoniae isolates, particularly those categorized as multidrug-resistant (MDR), XDR, and pandrug-resistant, are influenced not only by β-lactamase production, such as ESBL and metallo-β-lactamase, but also by the overexpression of efflux pump genes and reduced outer membrane permeability due to porin loss. These combined mechanisms significantly enhance bacterial survival against antibiotics, suggesting that effective control of AMR requires a multitargeted therapeutic approach that includes efflux pump inhibitors and strategies to restore membrane permeability. Carbapenem-resistant K. pneumoniae and multidrug resistance (MDR) have become serious global public health issues in recent years (Meletis, 2016). In 1983, the first instance of K. pneumoniae producing ESBL was identified (Rahim and Mohamed, 2014). The number of diseases caused by bacteria that produce ESBLs has increased globally in recent decades. In Indonesia, the percentage of Escherichia coli and K. pneumoniae infections caused by ESBL-producing bacteria is 29% and 36%, respectively (Sunarno et al., 2023). There are a number of traditional markets in Surabaya, where people actively buy and sell goods. Typically, egg traders are grouped with other traders rather than kept in a separate area. Cross-contamination between the sold products may result from this. One of the things that might lead to the growth of different microorganisms, including bacteria, that can harm the quality of eggs is cross contamination (Karanth et al., 2023). This study focused on the existence of K. pneumoniae as the main ESBL-producing bacteria that have MDR properties in chicken egg swabs in traditional markets, so that the characteristics of its resistance to antibiotics that have an impact on public health can be known. In addition, little data regarding the discovery of the blaSHV gene in ESBL-producing K. pneumoniae bacteria isolated from chicken egg swabs is the driving force behind this study. Moreover, the detection of this gene in food products, such as chicken eggs, indicates a potential transmission route of MDR bacteria to humans, which is a serious public health concern. Materials and MethodsResearch designThe study was conducted from November 2024 to January 2025. This research is classified as a cross-sectional study. Sampling of chicken egg shell swabs were sampled in traditional markets in Surabaya, Indonesia: Pacar Keling Market, Keputran, Manyar, Pucang, Sememi, Simo, Wonokromo, Wiyung, Pabean, and Kapas Krampung. The sampling locations were selected based on specific criteria, such as traditional markets with high visitor traffic, poor sanitation conditions, and a high number of egg vendors. The isolation and identification of bacteria and ESBL detection will be performed at the Veterinary Public Health Laboratory, Faculty of Veterinary Medicine, Airlangga University. Molecular detection of the blaSHV gene encoding ESBL in K. pneumoniae isolates using the PCR method will be carried out at the Institute of Tropical Disease Laboratory, Airlangga University. Sample collectionA total of 160 chicken egg shell swab samples, consisting of 80 layer chicken eggs and 80 free-range chicken eggs, were taken from 10 traditional markets in Surabaya using the criteria of looking dirty. The chicken egg shell swab technique was performed by wiping the chicken egg shell area with a sterile cotton swab moistened with physiological NaCl. The swab samples were then placed into labeled sample pots containing 1% Buffered Peptone Water as a transport medium and stored in a cooler box filled with ice packs at 4°C. Isolation and Identification of K. pneumoniaeThe egg shell swab results were mixed in a test tube containing 5 ml 1% Buffer Peptone Water (BPW) (Oxioid, UK) and then incubated at 37 °C for 24 hours. The sample on the BPW media was inoculated on the selective Mac Conkey Agar (MCA) media (HiMedia MH081) and incubated at 37°C for 24 hours. Klebsiella pneumoniae bacterial colonies on MCA media will appear pink, round, and large in size, and will appear mucoid. Colonies suspected of being K. pneumoniae bacteria were identified morphologically through Gram staining, then continued with biochemical tests using IMViC: Indol motility (SIM; HiMedia, India), Methyl Red (MR), and Voges Proskauer (VP) (MRVP; HiMedia, India), Citrate (SCA) (HiMedia, India), Triple Sugar Iron Agar (TSIA) (HiMedia, India), and Urease (Urea Agar) (HiMedia, India) (Dameanti et al., 2023). Antibiotic sensitivity testThe antibiotic resistance test of K. pneumoniae isolates was conducted using the Kirby-Bauer agar diffusion method. The antibiotics used were those that had been packaged in disc form. K. pneumoniae bacterial isolates were taken 1–2 colonies using an ose and then placed in 8 ml of physiological NaCl that had been tested for turbidity with the McFarland 0.5 standard (1–2 x 108 CFU/ml). The suspension was then taken as much as 0.2 ml of the isolate suspension was carefully smeared on the surface of Mueller–Hinton agar (MHA) and allowed to dry before placing the antibiotic disc. The media was divided into five parts, and then the discs were placed on the MHA’s media. Sensitivity testing was carried out using 5 antibiotics, namely Amoxicillin 25 µg (Oxoid CT0264B), Tetracycline 30 µg (Oxoid CT0054B), Streptomycin 10 µg (Oxoid CT0047B), Erythromycin 15 µg (Oxoid CT054B), and Ciprofloxacin 5 µg (Oxoid CT0425B). After planting the antibiotics, they were incubated at 37 °C for 24 hours. Interpretation of the inhibition zone diameter results was interpreted based on the Clinical and Laboratory Standards Institute (CLSI) standard (CLSI, 2023). If there are three or more types of antibiotics in different groups or classes that experience resistance, this can be called MDR (Effendi et al., 2021). Molecular detection of the blaSHV gene using PCRK. pneumoniae isolates that experienced MDR were then subjected to molecular-based testing using PCR to detect the presence of the blaSHV gene. K. pneumoniae isolates to be tested for PCR were re-cultured on Nutrient Agar (NA; Oxoid, UK) media and incubated at 37°C for 24 hours. DNA extraction procedure using a few colonies of each bacterial strain resuspended in 100 μl of distilled water and the boiling method according to the method described by Dameanti et al. (2023). The resulting DNA extraction samples were mixed with their respective DNA primers, Taq DNA polymerase enzyme, deoxyribonucleotide triphosphates, and buffer (Thermo Fisher Scientific Inc, and final volume reaction is 25 μl (Massachusetts, USA). The primers used in the PCR amplification process for the blaSHV gene are presented in Table 1. The amplification procedure began with an initial denaturation at 96°C for 5 minutes, followed by 35 cycles of 96°C for 1 minute (denaturation), 60°C for 1 minute (annealing), 72°C for 1 minute (extension), and a final extension at 72°C for 10 minutes (Dameanti et al. 2023). The PCR results were visualized by electrophoresis using 2% agarose gels (Invitrogen, USA) (Wibisono et al., 2020). The negative control in this test used nuclease-free water, whereas the positive control used K. pneumoniae. Ethical approvalNot needed for this study. ResultsThe results of the isolation and identification of K. pneumoniae bacteria from 160 samples of eggshell swabs of layer and free-range chickens showed that 41/160 (25.62%) were positive for K. pneumoniae bacteria, with the details of the eggshell swabs of layer chickens being positive as many as 28/41 (68.29%) and the results of the eggshell swabs of free-range chickens showing positive 13/41 (31.7%) can be seen in Table 2. The results of isolation on MCA media are shown in Figure 1A, and the results of the biochemical tests are shown in Figure 1B. Table 1. Primers for blaSHV gene detection.
Table 2. Isolation and identification results of K. pneumoniae bacteria.
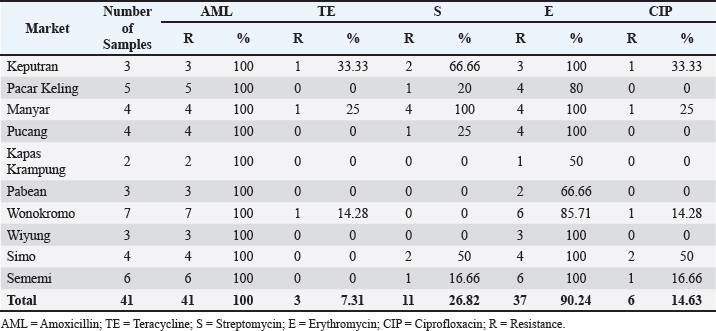
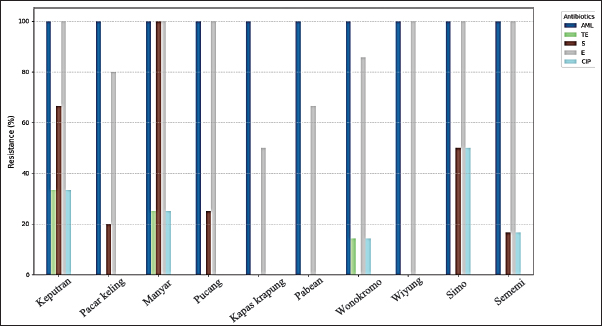
The highest isolation and identification results of K. pneumoniae bacteria were obtained from Wonokromo Market at a percentage of 43.75% (7/16). Furthermore, the Sememi market showed positive results of 37.50% (6/16), and Pacar Keling showed a percentage of 31.25% (5/16). Manyar, Pucang, and Simo markets showed 25.00% (4/16). Pabean, Wiyung, and Keputeran markets showed positive results of 18.75% (3/16). The lowest positive results were found for K. pneumoniae bacteria, 12.50% (2/16), from the traditional Kapas Krampung market. The results of antibiotic resistance tests on K. pneumoniae isolates from five different antibiotic groups obtained from eggshell swabs of layer and free-range chickens can be seen in Table 3, showing the highest incidence occurred in Amoxicillin antibiotics with a percentage of 100% (41/41) occurring in all traditional markets, then antibiotic resistance to Erythromycin 90.24% (37/41) with a distribution in the Keputran, Pucang, Manyar, Wiyung, Simo, Sememi markets 100%, followed by the Wonokromo market showing a percentage of 85.71% (6/7), Pacar Keling 80.00% (4/5), Pabean 66.66% (2/3), and Kapas Krampung with a percentage of 50% (1/2). The antibiotic resistance rate to streptomycin was 26.82% (11/41), with the highest incidence of resistance being Manyar market 100% (4/4), Keputran 100% (2/3), Simo 50.00% (2/4), Pucang 25.00% (1/4), Pacar Keling 20.00% (1/5), and no resistance was found in Kapas Krampung, Pabean, Wiyung, and Wonokromo markets. Antibiotic Ciprofloxacin was 14.63% (6/41). In the Simo market, it was 50% (2/4), Keputeran 33.33% (1/3), Manyar 25.00% (1/4), and Sememi 16.66% (1/6), and the lowest was Wonokromo 14.28% (1/7). The Pabean, Kapas Krampung, Pacar Keling, Pucang, and Wiyung markets were not found. Tetracycline antibiotic resistance was 7.31% (3/41), with the highest percentage coming from the Keputeran market (33.33% (1/3), followed by Manyar 25.00% (1/4), Wonokromo 14.28% (1/7), while Pacar Keling, Pucang, Pabean, Kapas Kerapung, Simo, Sememi, and Wiyung markets did not have Tetracycline antibiotic resistance (0%). Figure 2 shows the resistance patterns across different markets for comparison.
Fig. 1. Isolation and identification of the bacteria. (A) Colony of K. pneumoniae bacteria on MCA; (B) Biochemical test. Table 3. Identification of antibiotic resistance against K. pneumoniae isolates in layer and free-range chicken shells.
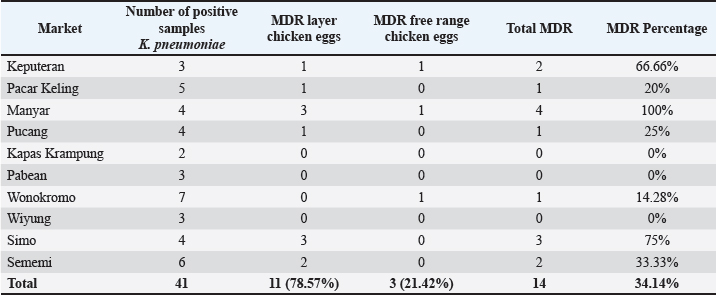
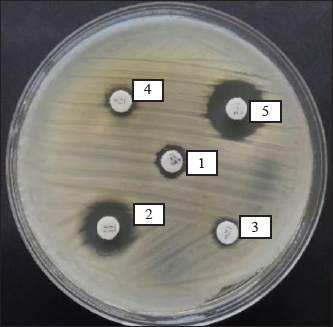
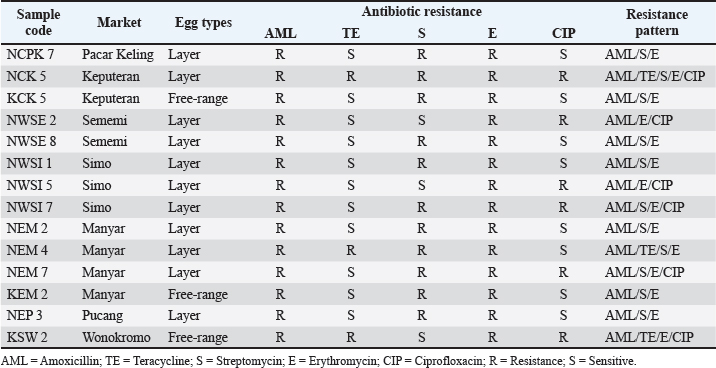
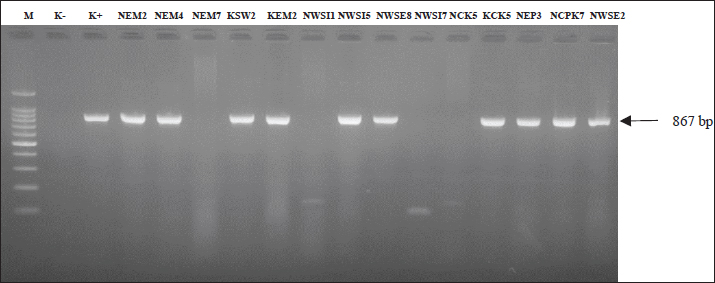
The results of antibiotic resistance tests on K. pneumoniae isolates from chicken egg shell swabs from traditional markets in Surabaya found that isolates that were resistant to 3 or more can be called MDR, 34.14% (14/41). The occurrence of MDR in K. pneumoniae isolates from chicken egg shell swabs was found with a percentage of 78.57% (11/14), whereas that in K. pneumoniae isolates from chicken egg shell swabs was found with a percentage of 21.42% (3/14). The highest percentage of MDR occurred in Manyar market, 100% (4/4), followed by Simo 75.00% (3/4), Keputran market 66.66% (2/3), Sememi 33.33%, Pucang 25.00% (1/4), Pacar Keling 20.00% (1/5), and the lowest percentage by Wonokromo market 14.28% (1/7) (Table 4). The MDR K. pneumoniae isolate is shown in Figure 3. The MDR isolate was specifically detected for the presence of the ESBL gene blaSHV at the genetic level through PCR testing. The resistance pattern of K. pneumoniae in traditional markets in Surabaya City showing the occurrence of MDR, is presented in Table 5. The occurrence of MDR in K. pneumoniae isolates with the highest to lowest resistance patterns included AML/S/E, there are 7 samples (7/14; 50%), consisting of NCPK code 7, KCK 5, NWSE 8, NWSI 1, NEM2, KEM2, and NEP3. Next, followed by AML/E/CIP of two samples (2/14; 14%) consisting of NWSE2 and NWSI5. Then, ATM/S/E/CIP of (2/14; 14.28%) was calculated. Furthermore, AML/TE/S/E (NEM4), AML/TE/E/CIP (KSW2), and AML/TE/S/E/CIP (NCK5) each resistance pattern has one sample (1/14; 7%). Based on the results of this study, there were 10 out of 14 K. pneumoniae isolates from MDR from eggshell swabs of Ras and Buras chickens showing a positive band at 867 base pairs (bp); thus, 71.42% (10/14) of K. pneumoniae isolates from MDR had the ESBL blaSHV gene (Fig. 4). The sample codes that were positive for the blaSHV gene included: NEM2, NEM4, KSW2, KEM2, NWSI5, NWSE8, KCK5, NEP3, NCPK7, and NWSE2. Negative sample codes included NEM7, NCK5, NWSI1, and NWSI7.
Fig. 2. Antibiotic resistance patterns of across different markets. Description: AML=Amoxicillin; TE=Tetracycline; S=Streptomycin; E=Erythromycin; CIP=Ciprofloxacin. Table 4. MDR incidence rate against K. pneumoniae bacteria.
DiscussionThe results of isolation and identification showed that 41 positive samples (25.62%) were identified as K. pneumoniae bacteria. Positive samples consisted of 28 swabs of layer chicken egg shells (68.29%) and 13 swabs of free-range chicken eggs (31.70%). The results of this study were higher than those previously conducted on egg samples from laying hen farms in Punjab, India; 11 (4.58%) were positive for K. pneumoniae from the litter cage system, and 7 (2.91%) from the battery cage system (Khehra, 2022). Research conducted by Permatasari et al. (2020) in Blitar, Indonesia, found K. pneumoniae contamination (3.87%) in cloacal swabs from laying hen farms. In another study conducted in Indonesia, the incidence of K. pneumoniae (7.8%) was recorded in chicken cloacal swabs (Hayati et al., 2019). Research by Aly et al. (2014) studied chick droppings and found K. pneumoniae contamination (10%) in their study. Differences in the research results may be influenced by the type of samples, such as feces, intestinal contents, or eggshells, which can affect bacterial isolation outcomes. Additionally, small sample sizes or inconsistent sampling techniques can lead to variations in the research results (Kahin et al., 2024).
Fig. 3. Results of the MDR K. pneumoniae bacteria test. Description: 1. Amoxicillin; 2. Tetracycline; 3. Streptomycin; 4. Erythromycin; 5. Ciprofloxacin. Table 5. The resistance pattern of MDR K. pneumoniae.
Fig. 4. PCR results of the blaSHV gene of K. pneumoniae isolates from multidrug-resistance strains; positive strain at band 867 bp. Description: M: Marker 100 bp; K-: Negative control; K+: Postive control Klebsiella pneumoniae is an opportunistic bacteria that is widely found in nature, such as soil, skin, intestines, and animal feces (Abbas et al., 2024). Klebsiella pneumoniae has also caused many cases of bloodstream infections or sepsis, pneumonia, and urinary tract infections (UTIs), and it is often found contaminating food and food ingredients, such as eggs (Riwu et al., 2022). The source of K. pneumoniae bacterial contamination in chicken eggs found in traditional markets is possible due to several factors. There are two types of contamination: vertical and horizontal. Infection of eggs as they develop in the oviduct is known as vertical contamination. In contrast to horizontal contamination, which is more frequent and refers to contamination of the eggshell surface after it has been deposited, this happens when laying chickens carry infections and transfer them to the eggs. This occurs when the eggshell comes into contact with a polluted surface (El Ftouhy et al., 2022). The high incidence of antibiotic resistance in this study indicates the high spread of antibiotic resistance in eggshells sold in traditional markets in Surabaya. On the basis of earlier claims that antimicrobial-resistant strains of K. pneumoniae have been detected in a variety of food sources across the globe, such as vegetables, fish, poultry, meat, eggs, milk, and dairy products (Junaid et al., 2022). In addition to direct contact between humans and infected animals or environments, or biological substances they produce such as blood, urine, feces, saliva, and semen, animals and plants that are used as human food sources can also serve as vectors for the transmission of antimicrobial-resistant bacteria and AMR genes to humans, resulting in contamination by resistant K. pneumoniae bacteria (Chang et al., 2015). The increase in resistance to common antibiotics is largely due to the uncontrolled use of antimicrobial agents to treat diseases and promote the growth of livestock; then, antibiotic residues can be spread into the environment, such as soil, water, and air (Nhung et al., 2017; Widodo et al., 2022; Khairullah et al., 2022). The mechanism of antibiotic resistance in bacteria involves various factors, including genetic mutations, horizontal genetic exchange through conjugation, transformation, and transduction, and selective pressure generated by repeated antibiotic exposure (Putri et al., 2023). Bacteria that are initially susceptible to certain antibiotics can develop defense mechanisms, such as enzymes that destroy antibiotics or change the molecular targets inhibited by antibiotics (Muteeb et al., 2023). In this study, Amoxicillin became the antibiotic with the highest resistance rate of 100% (41/41). Previous research data showed that K. pneumoniae isolates from bat feces showed a percentage of 100% resistance to Amoxicillin (Kinasih et al., 2024). The cause of the high level of Amoxicillin resistance is due to the ability of K. pneumoniae bacteria to produce beta-lactamase, an enzyme that can hydrolyze the beta-lactam ring in Amoxicillin, thereby deactivating its antibacterial activity. In addition to beta-lactamase production, other resistance mechanisms such as changes in penicillin-binding proteins, the presence of efflux pumps, and biofilm formation also contribute to the decreased effectiveness of Amoxicillin (Reygaert, 2018). The incidence of antibiotic resistance to erythromycin was 90.24% (37/41) ranks second highest. This result was higher than that of previous research on K. pneumoniae isolates (42.86%) (Permatasari et al., 2020). The effectiveness of Erythromycin is influenced by several factors, including the antibiotic concentration, environmental pH (its activity increases at alkaline pH), and bacterial susceptibility. Erythromycin resistance can arise as a result of changes to the 23S rRNA present in the 50S rRNA. Since erythromycin is unable to attach to ribosomes, bacteria are able to continue synthesizing proteins (Guo et al., 2015). External factors, such as inappropriate use of antibiotics and horizontal transfer of resistance genes through genetic elements, such as plasmids and transposons, accelerate the spread of Erythromycin resistance among bacterial populations. The incidence rate of streptomycin antibiotic resistance was 27% (11/41). Previous studies have shown that the level of K. pneumoniae resistance to streptomycin antibiotics is higher, namely 100% (10/10) (Kinasih et al., 2024). The results of the resistance test on the antibiotic streptomycin were 26.82% (11/41). This result was higher than that of previous studies on K. pneumoniae isolates (21.4%) (Permatasari et al., 2020). The mechanism of antibiotic resistance to streptomycin, which is a group of aminoglycoside antibiotics, is one of which is able to produce enzymes that modify antibiotics before they work on target cells. Antibiotic modification is a common strategy used by bacteria resistant to Streptomycin antibiotics. Enzymes that modify antibiotics include Acetyltransferase, Phosphotransferase, and adenyltransferase (Ramirez and Tolmasky, 2010). The efflux pump mechanism is a defense mechanism possessed by streptomycin-resistant bacteria (Krause et al., 2016). The incidence of Ciprofloxacin antibiotic resistance was 14.63% (6/41). The results of this study are lower than those of previous studies in which K. pneumoniae isolates were found to be 100% resistant (10/10) (Kinasih et al., 2024). Ciprofloxacin resistance is primarily caused by modifications to the target enzymes DNA gyrase and topoisomerase IV. Mutations in the Ciprofloxacin binding site reduce the affinity of the drug for these enzymes, thereby reducing its effectiveness (Laponogov et al., 2013). Changes in drug permeability also contribute to resistance. Bacteria can reduce ciprofloxacin penetration through modification of the cell membrane or reduction of porin channel activity, which prevents the drug from entering the cell (Hooper and Jacoby, 2015). Plasmid-mediated quinolone resistance is also a major factor in the spread of resistance. Plasmids carrying resistance genes can encode proteins that alter drug targets or enhance efflux pump mechanisms to remove Ciprofloxacin from the cell (Rodríguez-Martínez et al., 2016). The lowest resistance incidence was for tetracycline antibiotics [7.31% (3/41)]. Tetracycline is used to treat bacterial infections, such as the respiratory tract, urinary tract, skin, and pneumonia (Chopra and Roberts, 2001). Tetracycline resistance is primarily caused by ribosome defense mechanisms, enzymatic inactivation mechanisms, and efflux pumps. Eflux pumps are an example of a resistance mechanism that removes antibiotics from bacterial cells by exchanging protons as an energy source while maintaining low intracellular concentrations. Antimicrobials that enter the cell are pumped out before reaching the target. Eflux pumps occur when genes encode proteins. Eflux pumps are grouped into two, namely genes encoded on chromosomes and genes encoded on plasmids (tetK and tetL) (Dashtbani-Roozbehani and Brown, 2021). The data from this study show that K. pneumoniae bacterial isolates isolated from layer chicken eggs tend to be more resistant to antibiotics than native chicken eggs. This is due to several factors. First, layer chickens are often given antibiotics to prevent disease or improve their productivity, which can cause bacteria to become resistant to these drugs (Abreu et al., 2023). Second, layer chickens are kept in larger numbers in cages, so that the spread of bacteria is faster and makes antibiotic use higher, so that the transmission of resistance genes is high (Mehdi et al., 2018). In addition, the cleaning and sanitation processes on layer chicken egg farms are often not perfect, which allows for the development of resistant bacteria. On the other hand, layer chickens are kept in a more natural way and are rarely given antibiotics, so the possibility of contamination with resistant bacteria in layer chicken eggs is lower. The results of this study showed that 14 isolates experienced MDR from 41 K. pneumoniae isolates (34.14%) isolated from eggshell swabs of layer and free-range chickens. MDR is often associated with high mortality rates, prolonged disease, decreased treatment effectiveness, and increased medical costs that are very high (Tanwar et al., 2014). Information about K. pneumoniae that was isolated from Egyptian poultry farms. MDR Environmental samples from poultry farms contained K. pneumoniae strains in 16.7% of cases (Elmonir et al., 2021). In Malaysia, the frequency of MDR K. pneumoniae isolates from environmental samples was 20.2% (21/204), with 5/21 isolates accounting for 23.8% of the total (Veloo et al., 2022). There are two main mechanisms of antibiotic resistance in K. pneumoniae bacteria. The first mechanism is the expression of ESBL, which causes bacteria to become resistant to monobactams and cephalosporins. In the second mechanism, carbapenemases are expressed, which aids in the development of resistance to all β-lactams (Riwu et al., 2022). ESBL is produced by genes located on plasmids, so that bacteria that produce ESBL need to be monitored because these bacteria can easily move to other bacteria and often also carry resistance genes to other antibiotics such as aminoglycosides, quinolones, and co-trimoxazole, making it difficult to find alternative therapies (Jensen et al., 2006). Antibiotic resistance is influenced by the ESBL genes, which are encoded by plasmids in K. pneumoniae and include SHV, TEM, and CTX-M (Ahmed et al., 2014). The K. pneumoniae isolates that were multidrug resistant were then continued by detecting the presence of the ESBL-producing gene, namely blaSHV. PCR tests showed that there were 10/14 with a percentage of 71.42% of K. pneumoniae bacterial isolates from eggshell swabs having the blaSHV resistance gene. These results identified 10 of the 14 K. pneumoniae isolates that produced ESBL based on the detection of the blaSHV gene. One K. pneumoniae isolate did not harbor the blaSHV gene or possibly harbored another type of ESBL gene. According to previous research, SHV is the most prevalent kind of ESBL present in different Enterobacteriaceae, and it is typically observed in K. pneumoniae and E. coli (Ghafourian et al., 2015; Pehlivanoglu et al., 2016). The majority of K. pneumoniae strains, according to this study, produce blaSHV, although isolates of E. coli, Enterobacter species, and Klebsiella oxytoca were more likely to have blaTEM and blaCTX-M (Doi et al., 2017; Soekoyo et al., 2020). These findings raise serious concerns about the possibility of resistant genes spreading to people, animals, and the environment. In contrast to the CTX-M and TEM variations, the SHV enzyme has not expanded much, but in recent years, it has mostly been discovered in K. pneumoniae isolates with growing gene variations and in a variety of settings (Liakopoulos et al., 2016). In K. pneumoniae, the SHV ESBL enzyme is highly prevalent and accounts for 20% of plasmid-mediated penicillin group resistance (Shaikh et al., 2015). ESBL genes are quickly transferred from animals to humans or the other way around by mobile genetic elements such as transposons, DNA segment sequences, and integrons in bacteria (Jesumirhewe et al., 2020). These genetic factors can also spread resistance traits to other bacteria in the animal’s digestive tract. The bacteria then infect the soil and water surrounding the farm as they travel through waste made possible by inadequate sanitation and hygiene (Pradika et al., 2019). ESBL-producing bacteria have been found in produce, soil, and water near the farm and market in addition to the farm itself (Effendi et al., 2022). Contamination of egg products and other food ingredients can occur if the processing is not performed with good hygiene. This result demonstrates that ESBL bacteria cause foodborne illnesses and community infections in addition to nosocomial infections. ConclusionThis study identified the blaSHV gene in 71.42% (10/14) of K. pneumoniae MDR strains from eggshell swabs of both layer- and free-range chickens. This suggests that poultry products are a source of ESBL-producing bacteria transmission. These results can help veterinary authorities address the risk of AMR due to bacterial contamination of eggshells. Based on these findings, the government should create new policies for egg handling, including microbiological safety standards and sanitation checks before eggs are sold to the public to protect consumer health. These measures aim to improve public health and safety. Additionally, this study can raise awareness of food safety, especially for animal-based products, and encourage responsible antibiotic use. Future research should explore AMR in other foodborne pathogens and animal products and investigate how farm practices, antibiotic use, and environmental factors contribute to the spread of resistant bacteria. These efforts are crucial for developing strategies to tackle AMR globally through a One Health approach. AcknowledgmentsThe authors would like to thank the Universitas Airlangga. FundingThis study was partly supported by The International Research Consortium, Lembaga Penelitian dan Pengabdian Masyarakat, Universitas Airlangga, Surabaya, Indonesia, in the year 2024 with grant number: 171/UN3.LPPM/PT.01.03/2024. Author’s contributionsAAS, LRS, and ARK: Conceived, designed, and coordinated the study. PK, WT, and MHE: Designed data collection tools, supervised the field sample and data collection, and laboratory work, as well as data entry. IBM, KHPR, and SR: Validation, supervision, and formal analysis. SMY, AW, and BB: Contributed reagents, materials, and analysis tools. RZA and DAA: Statistical analysis and interpretation, and participated in the preparation of the manuscript. All authors have read, reviewed, and approved the final manuscript. Conflict of interestThe authors declare no conflict of interest. Data availabilityAll data are available in the manuscript. ReferencesAbbas, R., Chakkour, M., El Dine, H.Z., Obaseki, E.F., Obeid, S.T., Jezzini, A., Ghssein, G. and Ezzeddine, Z. 2024. General overview of Klebsiella pneumoniae: epidemiology and the role of siderophores in its pathogenicity. Biology (Basel) 13(2), 78. Abreu, R., Semedo-Lemsaddek, T., Cunha, E., Tavares, L. and Oliveira, M. 2023. Antimicrobial Drug Resistance in Poultry Production: Current Status and Innovative Strategies for Bacterial Control. Microorganisms 11(4), 953. Ahmed, D., Wahid, S.U., Sadique, T., Sultana, N., Islam, M., Halim, F., Islam, N. and Hossain, A. 2014. Recurrent urinary tract infection due to co-infection with extended spectrum β-lactamase-producer uropathogenic Escherichia coli and enteroaggregative E. coli. JMM Case Rep. 1(3), e001404. Aly, M.M., Khalil, S. and Metwaly, A. 2014. Isolation and molecular identification of Klebsiella microbe isolated from chicks. Alex. J. Vet. Sci. 43(1), 97–103. Ansharieta, R., Effendi, M.H. and Plumeriastuti, H. 2021. Genetic identification of shiga toxin encoding gene from cases of multidrug resistance (MDR) Escherichia coli isolated from raw milk. Trop. Anim. Sci. J. 44(1), 10–15. Benz, F., Huisman, J.S., Bakkeren, E., Herter, J.A., Stadler, T., Ackermann, M., Diard, M., Egli, A., Hall, A.R., Hardt, W.D. and Bonhoeffer, S. 2021. Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 15(3), 862–878. Bradley, M.E. and Scoular, S.K. 2021. Metastatic Klebsiella pneumoniae invasive liver abscess syndrome in Denver, Colorado. J. Pharm. Pract. 34(2), 332–336. Chang, Q., Wang, W., Regev-Yochay, G., Lipsitch, M. and Hanage, W.P. (2015), Antibiotics in agriculture and the risk to human health: how worried should we be?. Evol Appl. 8, 240–247. Chopra, I. and Roberts, M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65(2), 232–260 Clinical and Laboratory Standards Institute (CLSI). 2023. Performance standards for antimicrobial susceptibility testing. 33rd. ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. Dameanti, F.N.A.E.P., Yanestria, S.M., Widodo, A., Effendi, M.H., Plumeriastuti, H., Tyasningsih, W., Ugbo, E.N., Sutrisno, R. and Safri, M.A.A.S. 2023. Prevalence of multidrug resistance and extended-spectrum beta-lactamase-producing Klebsiella pneumoniae from dairy cattle farm wastewater in East Java Province, Indonesia. Int. J. One Health 9(2), 141–149. Dashtbani-Roozbehani, A. and Brown, M.H. 2021. Efflux pump mediated antimicrobial resistance by Staphylococci in health-related environments: challenges and the quest for inhibition. Antibiotics (Basel) 10(12), 1502. Dela, H., Egyir, B., Behene, E., Sulemana, H., Tagoe, R., Bentil, R., Bongo, R.N.A., Bonfoh, B., Zinsstag, J., Bimi, L. and Addo, K.K. 2023. Microbiological quality and antimicrobial resistance of Bacteria species recovered from ready-to-eat food, water samples, and palm swabs of food vendors in Accra, Ghana. Int. J. Food Microbiol. 396(1), 110195. Doi, Y., Iovleva, A. and Bonomo, R.A. 2017. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J. Travel Med. 24(suppl_1), S44–S51. Effendi, M.H., Harijani, N., Yanestria, S.M. and Hastutiek, P. 2018. Identification of Shiga toxin-producing Escherichia coli in raw milk samples from dairy cows in Surabaya, Indonesia. Philipp. J. Vet. Med. 55(SI), 109–114. Effendi, M.H., Hartadi, E.B., Witaningrum, A.M., Permatasari, D.A. and Ugbo, E.N. 2022. Molecular identification of blaTEM gene of extended-spectrum beta-lactamase-producing Escherichia coli from healthy pigs in Malang district, East Java, Indonesia. J. Adv. Vet. Anim. Res. 9(3), 447–452. Effendi, M.H., Tyasningsih, W., Yurianti, Y.A., Rahmahani, J., Harijani, N. and Plumeriastuti, H. 2021. Presence of multidrug resistance (MDR) and extended-spectrum beta-lactamase (ESBL) of Escherichia coli isolated from cloacal swabs of broilers in several wet markets in Surabaya, Indonesia. Biodiversitas 22(1), 304–310. El Ftouhy, F.Z., Nassik, S., Nacer, S., Kadiri, A., Charrat, N., Attrassi, K., Fagrach, A., Bahir, M.A., Derqaoui, S. and Hmyene, A. 2022. Bacteriological quality of table eggs in moroccan formal and informal sector. Int. J. Food Sci. 2022(1), 6223404. Elmonir, W., Abd El-Aziz, N.K., Tartor, Y.H., Moustafa, S.M., Remela, E.M.A., Eissa, R., Saad, H.A. and Tawab, A.A. 2021. Emergence of colistin and carbapenem resistance in extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from chickens and humans in Egypt. Biology (Basel) 10(5), 373. Ferdinand, A.S., McEwan, C., Lin, C., Betham, K., Kandan, K., Tamolsaian, G., Pugeva, B., McKenzie, J., Browning, G., Gilkerson, J., Coppo, M., James, R., Peel, T., Levy, S., Townell, N., Jenney, A., Stewardson, A., Cameron, D., Macintyre, A., Buising, K. and Howden, B.P. 2024. Development of a cross-sectoral antimicrobial resistance capability assessment framework. BMJ Glob. Health 9(1), e013280. Ghafourian, S., Sadeghifard, N., Soheili, S. and Sekawi, Z. 2015. Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr. Issues Mol. Biol. 17(1), 11–21. Guo, M.T., Yuan, Q.B. and Yang, J. 2015. Insights into the amplification of bacterial resistance to erythromycin in activated sludge. Chemosphere 136(1), 79–85. Hartantyo, S.H.P., Chau, M.L., Koh, T.H., Yap, M., Yi, T., Cao, D.Y.H., GutiÉrrez, R.A. and Ng, L.C. 2020. Foodborne Klebsiella pneumoniae: virulence potential, antibiotic resistance, and risks to food safety. J. Food Prot. 83(7), 1096–1103. Hayati, M., Indrawati, A., Mayasari, N.L.P.I., Istiyaningsih, I. and Atikah, N. 2019. Molecular detection of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates of chicken origin from East Java, Indonesia. Vet. World 12(4), 578–583. Hooper, D.C. and Jacoby, G.A. 2015. Mechanisms of drug resistance: quinolone resistance. Ann. N Y Acad. Sci. 1354(1), 12–31. Hussein, R.A., Al-Kubaisy, S.H. and Al-Ouqaili, M.T.S. 2024. The influence of efflux pump, outer membrane permeability and β-lactamase production on the resistance profile of multi, extensively and pandrug resistant Klebsiella pneumoniae. J. Infect. Public Health 17(11), 102544. Jensen, L.B., Hasman, H., Agersø, Y., Emborg, H.D. and Aarestrup, F.M. 2006. First description of an oxyimino-cephalosporin-resistant, ESBL-carrying Escherichia coli isolated from meat sold in Denmark. J. Antimicrob. Chemother. 57(4), 793–794. Jesumirhewe, C., Springer, B., Allerberger, F. and Ruppitsch, W. 2020. Whole genome sequencing of extended-spectrum β-lactamase genes in Enterobacteriaceae isolates from Nigeria. PLoS One 15(4), e0231146. Junaid, K., Ejaz, H., Younas, S., Alanazi, A., Yasmeen, H. and Rehman, A. 2022. Detection of Klebsiella pneumoniae antibiotic-resistant genes: an impending source of multidrug resistance dissemination through raw food. Saudi J. Biol. Sci. 29(5), 3347–3353. Kahin, M.A., Mohamed, A.H., Mohomed, A.A., Hassan, M.A., Gebremeskel, H.F. and Kebede, I.A. 2024. Occurrence, antibiotic resistance profiles and associated risk factors of Klebsiella pneumoniae in poultry farms in selected districts of Somalia Reginal State, Ethiopia. BMC Microbiol. 24(1), 137. Karanth, S., Feng, S., Patra, D. and Pradhan, A.K. 2023. Linking microbial contamination to food spoilage and food waste: the role of smart packaging, spoilage risk assessments, and date labeling. Front. Microbiol. 14(1), 1198124. Kendek, I.A., Putri, M.F.R., Wibisono, F.J., Effendi, M.H., Tyasningsih, W., Ugbo, E.N. and Agumah, N.B. 2024. Molecular detection of hlyF gene on multidrug resistance of avian pathogenic Escherichia coli isolated from ducks on wet markets of Surabaya, Indonesia. Biodiversitas 25(3), 1246–1253. Khairullah, A.R., Kurniawan, S.C., Sudjarwo, S.A., Effendi, M.H., Widodo, A., Moses, I.B., Hasib, A., Zahra, R.L.A., Gelolodo, M.A., Kurniawati, D.A., Riwu, K.H.P., Silaen, O.S.M., Afnani, D.A. and Ramandinianto, S.C. 2024. Kinship analysis of mecA gene of methicillin-resistant Staphylococcus aureus isolated from milk and risk factors from the farmers in Blitar, Indonesia. Vet. World 17(1), 216–225. Khairullah, A.R., Rehman, S., Sudjarwo, S.A., Effendi, M.H., Ramandinianto, S.C., Gelolodo, M.A., Widodo, A., Riwu, K.H.P. and Kurniawati, D.A. 2022. Detection of mecA gene and methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and risk factors from farms in Probolinggo, Indonesia. F1000Res. 11(1), 722. Khehra, A. 2022. Prevalence and characterization of antimicrobial resistance in Klebsiella pneumoniae and Enterococcus species from farmed poultry eggs, environment, and farm handlers [Master’s thesis]. Ludhiana, India: Guru Angad Dev Veterinary and Animal Sciences University. Kinasih, K.N., Mustika, Y.R., Puspitasari, Y., Tyasningsih, W., Agustin, A.L.D., Kurniawan, S.C., Hasib, A., Waruwu, Y.K.K. and Silaen, O.S.M. 2024. Molecular detection of Klebsiella pneumoniae producing extended-spectrum beta-lactamase isolated from bat feces from the Tanjung Ringgit bat cave, Lombok Island, Indonesia. Int. J. One Health 10(1), 133–140. Krause, K.M., Serio, A.W., Kane, T.R. and Connolly, L.E. 2016. Aminoglycosides: an overview. Cold Spring Harb. Perspect. Med. 6(6), a027029. Kuang, H., Yang, F., Zhang, Y., Wang, T. and Chen, G. 2018. The impact of egg nutrient composition and its consumption on cholesterol homeostasis. Cholesterol 2018(1), 6303810. Laponogov, I., Veselkov, D.A., Crevel, I.M., Pan, X.S., Fisher, L.M. and Sanderson, M.R. 2013. Structure of an ‘open’ clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport. Nucleic Acids Res. 41(21), 9911–9923. Liakopoulos, A., Mevius, D. and Ceccarelli, D. 2016. A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front. Microbiol. 7(1), 1374 Mehdi, Y., Létourneau-Montminy, M.P., Gaucher, M.L., Chorfi, Y., Suresh, G., Rouissi, T., Brar, S.K., Côté, C., Ramirez, A.A. and Godbout, S. 2018. Use of antibiotics in broiler production: global impacts and alternatives. Anim. Nutr. 4(2), 170–178. Meletis, G. 2016. Carbapenem resistance: overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 3(1), 15–21. Muteeb, G., Rehman, M.T., Shahwan, M. and Aatif, M. 2023. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: a narrative review. Pharmaceuticals (Basel) 16(11), 1615. Nhung, N.T., Chansiripornchai, N. and Carrique-Mas, J.J. 2017. Antimicrobial resistance in bacterial poultry pathogens: a review. Front. Vet. Sci. 4(1), 126. Owaid, H.A. and Al-Ouqaili, M.T.S. 2025. Whole genome sequencing insights on extensive drug resistant Klebsiella pneumoniae and Pseudomonas aeruginosa traumatic infection. Pak. J. Biol. Sci., 28(2), 78–87. Pehlivanoglu, F., Turutoglu, H., Ozturk, D. and Yardimci, H. 2016. Molecular characterization of ESBL-producing Escherichia coli isolated from healthy cattle and sheep. Acta Vet. 66(4), 520–533. Permatasari, D.A., Witaningrum, A.M., Wibisono, F.J. and Effendi, M.H. 2020. Detection and prevalence of multidrug-resistant Klebsiella pneumoniae strains isolated from poultry farms in Blitar, Indonesia. Biodiversitas 21(10), 4642–4647. Pourhossein, Z., Asadpour, L., Habibollahi, H. and Shafighi, S.T. 2020. Antimicrobial resistance in fecal Escherichia coli isolated from poultry chicks in northern Iran. Gene Rep. 21(1), 100926. Pradika, A.Y., Chusniati, S., Purnama, M.T.E., Effendi, M.H., Yudhana, A. and Wibawati, P.A. 2019. Total test of Escherichia coli on fresh cow milk at dairy farmer cooperative (KPSP) Karyo Ngremboko Purwoharjo Banyuwangi. J. Med. Vet. 2(1), 1–6. Putri, M.F.R., Kendek, I.A., Wibisono, F.J., Effendi, M.H., Rahardjo, D., Tyasningsih, W. and Ugbo, E.N. 2023. Molecular detection of iron gene on multidrug resistant avian fecal Escherichia coli isolated from broiler on traditional markets, Surabaya, Indonesia. Biodiversitas 24(12), 6454–6460. Rahim, K.A.A.A. and Mohamed, A.M.A. 2014. Prevalence of extended spectrum β-lactamase-producing Klebsiella pneumoniae in clinical isolates. Jundishapur. J. Microbiol. 7(11), e17114. Ramirez, M.S. and Tolmasky, M.E. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13(6), 151–171. Reygaert, W.C. 2018. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 4(3), 482–501. Riwu, K.H.P., Effendi, M.H., Rantam, F.A., Khairullah, A.R. and Widodo, A. 2022. A review: Virulence factors of Klebsiella pneumoniae as emerging infection on the food chain. Vet. World 15(9), 2172–2179. Rodríguez-Martínez, J.M., Machuca, J., Cano, M.E., Calvo, J., Martínez-Martínez, L. and Pascual, A. 2016. Plasmid-mediated quinolone resistance: two decades on. Drug Resist. Updat. 29(1), 13–29. Salam, M.A., Al-Amin, M.Y., Salam, M.T., Pawar, J.S., Akhter, N., Rabaan, A.A. and Alqumber, M.A.A. 2023. Antimicrobial resistance: a growing serious threat for global public health. Healthcare (Basel) 11(13), 1946. Shaikh, S., Fatima, J., Shakil, S., Rizvi, S.M. and Kamal, M.A. 2015. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J. Biol. Sci. 22(1), 90–101. Soekoyo, A.R., Sulistiawati, S., Setyorini, W. and Kuntaman, K. 2020. The epidemiological pattern and risk factor of ESBL (Extended Spectrum β-Lactamase) producing Enterobacteriaceae in gut bacterial flora of dairy cows and people surrounding in rural area, Indonesia. Indones. J. Trop. Infect. Dis. 8(3), 144–151. Sunarno, S., Puspandari, N., Fitriana, F., Nikmah, U.A., Idrus, H.H. and Panjaitan, N.S.D. 2023. Extended spectrum beta lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae in Indonesia and South East Asian countries: GLASS Data 2018. AIMS Microbiol. 9(2), 218–227. Tanwar, J., Das, S., Fatima, Z. and Hameed, S. 2014. Multidrug resistance: an emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014(1), 541340. Veloo, Y., Thahir, S.S.A., Rajendiran, S., Hock, L.K., Ahmad, N., Muthu, V. and Shaharudin, R. 2022. Multidrug-resistant gram-negative bacteria and extended-spectrum β-lactamase-producing Klebsiella pneumoniae from the poultry farm environment. Microbiol. Spectr. 10(3), e0269421. Wibisono, F.J., Sumiarto, B., Untari, T., Effendi, M.H., Permatasari, D.A. and Witaningrum, A.M. 2020. CTX gene of Extended Spectrum Beta-Lactamase (ESBL) producing Escherichia coli on broilers in Blitar, Indonesia correspondence. Sys. Rev. Pharm. 11(7), 396–403. Widodo, A., Lamid, M., Effendi, M.H., Khailrullah, A.R., Kurniawan, S.C., Silaen, O.S.M., Riwu, K.H.P., Yustinasari, L.R., Afnani, D.A., Dameanti, F.N.A.E.P. and Ramandinianto, S.C. 2023. Antimicrobial resistance characteristics of multidrug resistance and extended-spectrum beta-lactamase producing Escherichia coli from several dairy farms in Probolinggo, Indonesia. Biodiversitas 24(1), 215–221. Widodo, A., Lamid, M., Effendi, M.H., Khairullah, A.R., Riwu, K.H.P., Yustinasari, L.R., Kurniawan, S.C., Ansori, A.N.M., Silaen, O.S.M. and Dameanti, F.N.A.E.P. 2022. Antibiotic sensitivity profile of multidrug-resistant (MDR) Escherichia coli isolated from dairy cow’s milk in Probolinggo, Indonesia. Biodiversitas 23(10), 4971–4976. Yuwono, Y. 2013. Identification of the SHV gene in Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs). Syifa’ Med. J. Med. Health. 4(1), 9–15. | ||
| How to Cite this Article |
| Pubmed Style Sulaiman AA, Effendi MH, Srimaryanto LR, Khairullah AR, Kurniasari P, Tyasningsih W, Moses IB, Ahmad RZ, Rehman S, Budiastuti B, Afnani DA, Yanestria SM, Riwu KHP, Widodo A. Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya. Open Vet. J.. 2025; 15(5): 2193-2205. doi:10.5455/OVJ.2025.v15.i5.37 Web Style Sulaiman AA, Effendi MH, Srimaryanto LR, Khairullah AR, Kurniasari P, Tyasningsih W, Moses IB, Ahmad RZ, Rehman S, Budiastuti B, Afnani DA, Yanestria SM, Riwu KHP, Widodo A. Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya. https://www.openveterinaryjournal.com/?mno=244245 [Access: January 24, 2026]. doi:10.5455/OVJ.2025.v15.i5.37 AMA (American Medical Association) Style Sulaiman AA, Effendi MH, Srimaryanto LR, Khairullah AR, Kurniasari P, Tyasningsih W, Moses IB, Ahmad RZ, Rehman S, Budiastuti B, Afnani DA, Yanestria SM, Riwu KHP, Widodo A. Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya. Open Vet. J.. 2025; 15(5): 2193-2205. doi:10.5455/OVJ.2025.v15.i5.37 Vancouver/ICMJE Style Sulaiman AA, Effendi MH, Srimaryanto LR, Khairullah AR, Kurniasari P, Tyasningsih W, Moses IB, Ahmad RZ, Rehman S, Budiastuti B, Afnani DA, Yanestria SM, Riwu KHP, Widodo A. Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya. Open Vet. J.. (2025), [cited January 24, 2026]; 15(5): 2193-2205. doi:10.5455/OVJ.2025.v15.i5.37 Harvard Style Sulaiman, A. A., Effendi, . M. H., Srimaryanto, . L. R., Khairullah, . A. R., Kurniasari, . P., Tyasningsih, . W., Moses, . I. B., Ahmad, . R. Z., Rehman, . S., Budiastuti, . B., Afnani, . D. A., Yanestria, . S. M., Riwu, . K. H. P. & Widodo, . A. (2025) Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya. Open Vet. J., 15 (5), 2193-2205. doi:10.5455/OVJ.2025.v15.i5.37 Turabian Style Sulaiman, Azhari Athaillah, Mustofa Helmi Effendi, Leonardo Reza Srimaryanto, Aswin Rafif Khairullah, Pitaloka Kurniasari, Wiwiek Tyasningsih, Ikechukwu Benjamin Moses, Riza Zainuddin Ahmad, Saifur Rehman, Budiastuti Budiastuti, Daniah Ashri Afnani, Sheila Marty Yanestria, Katty Hendriana Priscilia Riwu, and Agus Widodo. 2025. Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya. Open Veterinary Journal, 15 (5), 2193-2205. doi:10.5455/OVJ.2025.v15.i5.37 Chicago Style Sulaiman, Azhari Athaillah, Mustofa Helmi Effendi, Leonardo Reza Srimaryanto, Aswin Rafif Khairullah, Pitaloka Kurniasari, Wiwiek Tyasningsih, Ikechukwu Benjamin Moses, Riza Zainuddin Ahmad, Saifur Rehman, Budiastuti Budiastuti, Daniah Ashri Afnani, Sheila Marty Yanestria, Katty Hendriana Priscilia Riwu, and Agus Widodo. "Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya." Open Veterinary Journal 15 (2025), 2193-2205. doi:10.5455/OVJ.2025.v15.i5.37 MLA (The Modern Language Association) Style Sulaiman, Azhari Athaillah, Mustofa Helmi Effendi, Leonardo Reza Srimaryanto, Aswin Rafif Khairullah, Pitaloka Kurniasari, Wiwiek Tyasningsih, Ikechukwu Benjamin Moses, Riza Zainuddin Ahmad, Saifur Rehman, Budiastuti Budiastuti, Daniah Ashri Afnani, Sheila Marty Yanestria, Katty Hendriana Priscilia Riwu, and Agus Widodo. "Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya." Open Veterinary Journal 15.5 (2025), 2193-2205. Print. doi:10.5455/OVJ.2025.v15.i5.37 APA (American Psychological Association) Style Sulaiman, A. A., Effendi, . M. H., Srimaryanto, . L. R., Khairullah, . A. R., Kurniasari, . P., Tyasningsih, . W., Moses, . I. B., Ahmad, . R. Z., Rehman, . S., Budiastuti, . B., Afnani, . D. A., Yanestria, . S. M., Riwu, . K. H. P. & Widodo, . A. (2025) Molecular detection of blaSHV gene in multidrug resistance of Klebsiella pneumoniae isolated from chicken egg shell swab from a traditional market in Surabaya. Open Veterinary Journal, 15 (5), 2193-2205. doi:10.5455/OVJ.2025.v15.i5.37 |