| Research Article | ||
Open Vet. J.. 2025; 15(5): 2094-2102 Open Veterinary Journal, (2025), Vol. 15(05): 2094-2102 Research Article Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in EgyptYumna Elsobky1, Ibrahim M. Rabah2, Walid S. Mousa3, Khaled Sabbah3, Mohamed A. Nayel3, Ahmed M. Elsify3, Asmaa A. Elgendy4, Ahmed A. Zaghawa3, Akram A. Salam3, Ashraf M. Abu-Seida5*, Abdulrahman Abdulkarim6 and Mohamed M. Elkamshishi21Department of Hygiene and Zoonosis, Faculty of Veterinary Medicine, University of Sadat City, Sadat City, Egypt 2Department of Animal Hygiene and Zoonoses, Faculty of Veterinary Medicine, Matrouh University, Matrouh, Egypt 3Department of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City, Sadat City, Egypt 4Department of Bacteriology, Immunology and Mycology, Animal Research Institute (Shebin El kom Branch), Agriculture Research Center, Giza, Egypt 5Department of Surgery, Anesthesiology and Radiology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt 6Faculty of Veterinary Medicine, Omar Almukhtar University, Bayda, Libya *Corresponding Author: Ashraf M. Abu-Seida. Department of Surgery, Anesthesiology, and Radiology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt. Email: ashrafseida [at] cu.edu.eg Submitted: 27/01/2025 Revised: 13/04/2025 Accepted: 24/04/2025 Published: 31/05/2025 © 2025 Open Veterinary Journal
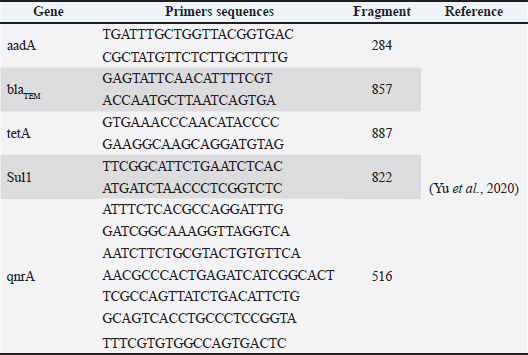
ABSTRACTBackground: Mastitis is a common disease that affects the dairy sector globally because it not only impacts animal welfare but can also lead to significant financial losses. Aim: This study examined the phenotypic and genotypic profiles of the multidrug-resistant (MDR) Escherichia coli (E. coli) strains that were isolated from mastitic cows in Egypt to detect their pattern of antibiotic resistance. Methods: Four hundred native breed lactating cows were evaluated to identify clinical and subclinical mastitis. A total of 100 mastitic milk samples (64 from clinical mastitis and 36 from subclinical mastitis) were collected for phenotypic isolation and identification of coliform bacteria. Escherichia coli isolates were identified through their morphological features, Gram staining, and biochemical tests. The identified E. coli strains were examined against various antibiotics using disk diffusion methods. All E. coli strains were analyzed for the antibiotic resistance genes Streptomycin (aadA), blaTEM, Tetracycline (tetA), Sulfonamides, and qnrA using PCR. Results: Among 400 examined dairy cows, the prevalences of clinical and subclinical mastitis were 16% and 9%, respectively. Bacteriological isolation of coliform bacteria from mastitic milk samples revealed that E. coli was the most prevalent bacterium. Among 10 isolates of biochemically verified E. coli strains, 8 (80%) were MDR across 6 distinct classes of antibiotics. All recovered E. coli strains exhibited higher resistance to Amoxicillin, Cefotaxime, Sulphamethaxzole/Trimethoprim, and Tetracycline. High susceptibility was noticed to Ciprofloxaccin, Amoxicillin+Clavulinic, Streptomycin, Gentamicin, Chloramphenicol, and Colistin. The blaTEM gene was among the most common antibiotic resistance genes found in E. coli isolates (100%). Furthermore, the genotypes encoding resistance to tetA, aadA, and Sulfonamides were 50%, 40%, and 50%, respectively. Conclusion: MDR pathogenic E. coli strains are common in mastitic dairy cows in Egypt, and preventive actions must be implemented to avoid serious public health concerns. Keywords: Antibiogram, Coliform bacteria, Dairy cows, Mastitis, Milk, Resistance genes. IntroductionMastitis is an inflammation of the mammary glands, which is usually caused by bacterial infections. However, fungal, viral, and environmental factors can also play a role in the development of this disease. It is one of the most prevalent and economically significant diseases affecting dairy herds worldwide (Abebe et al., 2016; Stanek et al., 2024). In addition to having an adverse effect on animal welfare, mastitis causes large financial losses because of decreased milk production, altered milk quality, medical expenses, and sometimes the need to cull afflicted animals (Cheng and Han, 2020). There are two primary types of mastitis: clinical and subclinical. In addition to aberrant milk properties, clinical mastitis is characterized by swelling, redness, hotness, and pain in affected udders. Somatic cell count and milk culture can be used to identify subclinical mastitis, which is more subtle and does not present with clinical symptoms (Cheng and Han, 2020). Mastitis in dairy cattle is linked to a number of variable risk factors, including the animal breed, season, somatic cell count, lactation stage, parity, and history of mastitis in both primiparous and multiparous cows (Oliveira et al., 2015). Cattle mastitis can be effectively managed and prevented by combining appropriate hygiene, prompt treatment, nutritional support, and good milking practices (Tommasoni et al., 2023). Antibiotic therapy has been the most widely used treatment for mastitis in cattle (de Souza et al., 2024). A variety of antibiotic types showed resistance in a number of bacterial species. Therefore, the rise of antibiotic-resistant bacterial strains has hindered the use of antibiotic-based mastitis therapy (Ameen et al., 2019). The majority of coliform bacterial isolates obtained from dairy animals showed a pattern of increased antibiotic resistance (Kateete et al., 2012). Multidrug-resistant (MDR) Escherichia coli (E. coli) has become a significant issue in the treatment of mastitis in cattle. Escherichia coli is a frequent environmental pathogen responsible for mastitis in dairy cattle. The rising occurrence of MDR strains complicates mastitis treatment protocols in cattle because these strains are resistant to multiple antibiotics frequently used in veterinary medicine (Pascu et al., 2022). Several factors contribute to multidrug resistance. These elements encompass the overuse or misuse of antibiotics, unsanitary practices on the farm, and the inherent ability of bacteria to gain and exchange resistance genes (Endale et al., 2023). Understanding the dynamics of MDR bacteria is essential for enhancing mastitis management and reducing the risks of antimicrobial resistance in both veterinary and human health care. Thus, many studies are essential to identify new biocontrol agents, especially in various developing nations, including Egypt. This research deals with isolation and genotypic characteristics of MDR E. coli strains obtained from mastitic cows in Egypt and examination of their antibiotic susceptibility patterns. Materials and MethodsAnimal examination and sample collectionThis research was conducted over the timeframe from October 2022 to June 2023. According to the cardinal signs of inflammation in the udder and systemic response, 400 native breed lactating cows (Mean age: 3.5 ± 0.8 years) were evaluated for identification of clinical mastitis following Bartlett et al. (2001). Additionally, apparently normal milk samples were collected from all apparently healthy cows and subjected to CMT for detection of subclinical mastitis as described by Balamurugan and Ranjith (2018). Phenotypic isolation and identification of Coliform bacteriaA total of 100 mastitic milk samples (64 from clinical mastitis and 36 from subclinical mastitis) were collected in a sterile manner and transported immediately under cool conditions, as mentioned by Cabral et al. (2015), for the isolation of coliform bacteria. Initially, the samples were inoculated into tryptose soy broth (mTSB-Difco La Jolla, CA, USA) and incubated for a duration of 12 hours at 37°C. The specimens were subcultured on a selective growth medium (MacConkey agar, MAC; Difco, La Jolla, CA, USA) and incubated at 37°C for 24–48 hours. Eosin methylene blue (EMB; Difco) medium was used to subculture the pink colonies that appeared on the plate with fermented lactose. Colony formations with a metallic green color were identified as E. coli. All isolates were identified as E. coli via Gram staining and their morphological features. Moreover, E. coli was verified through biochemical tests (Indole, citrate utilization, Voges–Proskauer, methyl red, triple sugar iron agar, and urease assays), as recommended by Cowan et al. (2015). Antibiogram patterns of E. coli isolatesThe Kirby–Bauer disk diffusion method from the Clinical and Laboratory Standards Institute was utilized for evaluation of the identified E. coli strains against various antibiotics (CLSI, 2018). To obtain turbidity equivalent to a 0.5 McFarland standard (1.5 × 108 colony-forming units (CFUs) ml−1), a suspension of the organism was prepared. After screening the isolates, the results were interpreted using the criteria specified by the CLSI. Oxoid, an upcoming generation of commercial antibiotic discs, was used. The following commercial antibiotic discs (Oxoid) were used: Amoxicillin (AX) 25 mcg, Amoxicillin/Clavulanic (AMC) 30 µg, Cefotaxime (CTX) 30 mg, Gentamicin (CN) 10 mcg, Tetracycline (TE) 30 mcg, Ciprofloxacin (CIP) 5 mcg, Streptomycin (S) 10 mg, Trimethoprim–Sulfamethoxazole (SXT) 125/23.75 mcg, Chloramphenicol (C) 30 mcg, and Colistin (CT) 10 µg. The outcomes were noted as susceptible, intermediate, or resistant, and they were assessed by the CLSI (2018). Molecular detection of an E. coli antibiotic resistance gene in mastitic cowsFollowing the manufacturer’s guidelines, QIAamp DNA Mini test kits (Qiagen, GmbH, Germany) were used for DNA extraction. All E. coli strains were analyzed for the antibiotic resistance genes Streptomycin (aadA), blaTEM, Tetracycline (tetA), Sulfonamides (sul1), and qnrA using PCR. The target genes of the E. coli isolates were identified using several PCR methods. Table 1 lists the amplified products, base sequences, and primers used. A PCR thermocycler (Applied Biosystems 2720) was used in all experiments. A total of 25 µl of DNA was amplified and comprised 12.5 µl of PCR Master Mix, 1 µl of each primer at a concentration of 20 pmol, 4.5 µl of filtered water, and 6 µl of the DNA template. Gel electrophoresis (1.5% agarose) was conducted on the amplified products, which were then stained with Ethidium bromide and captured using a UV transilluminator. Table 1. Lists of primers used, base sequences, and predicted sizes of amplified products.
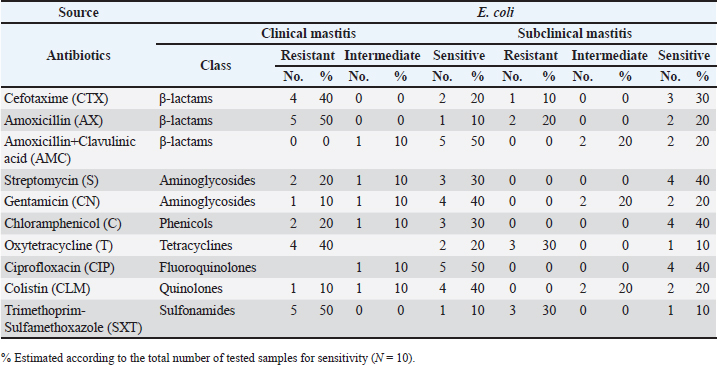
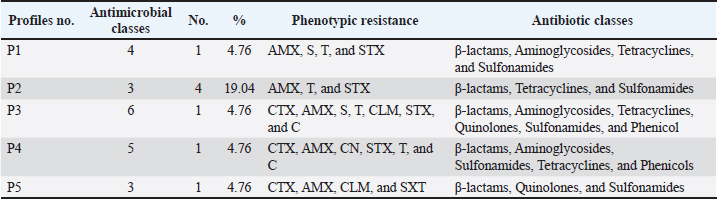
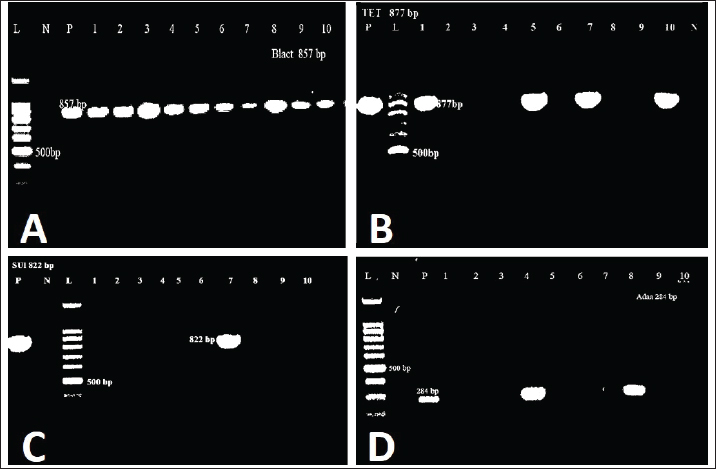
Statistical analysisData were analyzed using IBM SPSS Statistics 25 (SPSS Inc., Chicago, IL). The prevalence rates were calculated for all data as the number of infected individuals divided by the total number of individuals examined and multiplied by 100. Ethical approvalThis research received approval from the Ethical Committee at the Faculty of Veterinary Medicine, University of Sadat City, Egypt (Approval no. VUSC-014-2-20). ResultsPrevalence of clinical and subclinical mastitis in the examined cowsAmong 400 lactating native breed cows, the prevalence of clinical and subclinical mastitis was 16% (64/400) and 9% (36/400), respectively. The clinical assessment of cows suffering from clinical mastitis (n=64) showed significant udder inflammation, along with the presence of clots, flakes, and blood in the milk, frequently accompanied by systemic reactions such as fever, depression, and food sensitivity. Subclinical mastitis cases (n=36) exhibited varying levels of positive precipitation and gel formation in the CMT. Identification of E. coli isolates and antibiotic resistance patternsThe bacteriological isolation of coliform bacteria in the examined 100 mastitic milk samples revealed their presence in 53% (53/100) of the mastitic cases. Escherichia coli was the most common coliform bacterium, accounting for 15.62% of clinical mastitis cases (10/64) and 19.44% of subclinical mastitis cases (7/36). Additionally, other coliform bacteria were isolated from the examined 100 mastitic milk samples including Klebsiella spp. (9.76%), Citrobacter (6.1%), and Enterobacter aerogenes (3.66%). All identified E. coli strains showed a significant level of resistance to Amoxicillin, Cefotaxime, Sulphamethaxzole/Trimethoprim, and Tetracycline at rates of 71.4%, 71.4%, 66.6%, and 52.4%, respectively. In the meantime, susceptibility was noted to Ciprofloxacin, Amoxicillin+Clavulinic, Streptomycin, Gentamicin, Chloramphenicol, and Colistin, with rates of 95.2%, 76.2%, 66.7%, 61.9%, 52.4%, and 47.6%, respectively, as displayed in Table 2. Among 10 isolates of biochemically verified E. coli strains, 8 (80%) were MDR across 6 distinct classes of antibiotics, including β-lactams, Aminoglycosides, Macrolides, Tetracyclines, Quinolones, Sulfonamides, and phenicol Table 3. The studied isolates exhibited 6 distinct phenotypes. PCR results of E. coli strains’ antibiotic resistance genesPCR assays were employed to examine specific antibiotic resistance genes in all E. coli strains, as demonstrated in Figure 1A–D. All analyzed E. coli isolates displayed complete resistance to blaTEM based on molecular screening for antibiotic resistance genes. Moreover, the main resistance genes identified in the isolated E. coli strains were those conferring resistance to tetA, aadA, and sul1, with corresponding incidence rates of 50%, 40%, and 50%, respectively. In contrast, the qurA gene was absent in all of the samples. Table 2. Antibiogram patterns of E. coli strains isolated from clinically and subclinically mastitis dairy cows.
Table 3. Phenotypic profile of multidrug-resistant E. coli strains from the examined cows with mastitis.
DiscussionIn dairy farms, one of the primary causes of rising antimicrobial-resistant bacteria is mastitis, which arises from the widespread use of antibiotics (Ismail and Abutarbush, 2020). Numerous major bacterial species such as Coagulase-negative Staphylococci, Enterococci, Streptococci, and E. coli are linked to clinical bovine mastitis, with the rise of MDR bacterial strains (Kateete et al., 2013). The present research indicated that the prevalence rates of clinical and subclinical mastitis were 16% and 9%, respectively. In this context, numerous earlier studies have been carried out in Egypt by various researchers, including Mousa et al. (2015), who noted that the prevalence of both clinical and subclinical mastitis was 20.5% and 32%, respectively, and ElFaramaway et al. (2019), who found clinical mastitis in 30% of 400 cows assessed. Moreover, Ahmed et al. (2018), Abed et al. (2021), and Hussein et al. (2022) found a greater prevalence of mastitis in dairy cattle in Egypt, 49.9%, 46%, and 30.23%, respectively. The differences in mastitis prevalence across various studies may be due to the number and location of the cows examined, the samples taken, management practices, climatic variations across different areas, and the quality of veterinary care provided for the control and treatment of mastitis. Globally, East Africa had the significantly highest pooled prevalence estimate of subclinical mastitis (67.7%), followed by West Africa (50.5%), and the lowest was in North Africa (40.3%) (Khasapane et al., 2023). In South-Asian low-income countries like Bangladesh, India, Pakistan, and Sri Lanka, the prevalence of subclinical mastitis ranged from approximately 20% to approximately 80%, but the average prevalence across all studies was high (50%) (Bari et al., 2022). The overall mean prevalence of subclinical mastitis in a large population of Brazilian dairy herds (1809 herds) was 46.4% (Busanello et al., 2017).
Fig. 1. Agarose gel electrophoresis 1.5% showing (A): the blaTEM gene of E. coli at (857) bp: Lane M: 100 bp DNA ladder, +ve: Positive control, −ve: Negative control, and Lane 1–10 showing positive results for blaTEM gene in all isolates, (B): the tetA gene of E. coli at (877) bp: Lane M: 100 bp DNA ladder, +ve: Positive control, −ve: Negative control, Lanes 1, 5, 7, and 10 were positive for tetA gene and Lanes 2–4, 6, 8, and 9 were negative for tetA gene, (C): the sul1 gene of E. coli at (822) bp: Lane M: 100 bp DNA ladder, +ve: Positive control, −ve: Negative control, Lanes 7 and10 were positive for sul1 gene, and (D): the aadA gene of E. coli at (284) bp: Lane M: 100 bp DNA ladder, +ve: Positive control, −ve: Negative control, Lanes 1, 2, 4, and 5 were positive for aadA gene and Lane 3 was negative for the aadA gene. The cows affected by clinical mastitis exhibited significant udder inflammation and clots, flakes, and blood in milk, along with systemic reactions in most instances, whereas subclinical cases displayed varying levels of positive precipitation and gel formation in CMT. These results are in agreement with those of an earlier study (Elbably and Asmaa, 2013). In the present study, the microbiological isolation of coliform bacteria from mastitic milk samples revealed that E. coli was the most common bacterium. Comparable results were noted in previous studies (Elbably and Asmaa, 2013; Ahmed et al., 2018; El-Mohandes et al., 2022). In China, Yu et al. (2020) and Xu et al. (2023) also demonstrated that E. coli is a common mastitis pathogen in dairy farms. Likewise, Goulart and Mellata (2022) reported that pathogenic E. coli constitutes a major bacterial agent of acute clinical mastitis in dairy cattle. In contrast, Abegewi et al. (2022) reported that Enterobacter cloacae was the most prevalent bacteria causing mastitis in dairy cattle (12.6%). Antibiotic treatment is an important approach for managing various bacterial infections in both animals and humans (Adzitey et al., 2015). In the global dairy cattle population, antibiotic-resistant pathogens have emerged (Ameen et al., 2019). Specifically, E. coli strains that develop high resistance to antimicrobial drugs used in human and veterinary medicine, as reported by the Food and Drug Administration (2011), have been identified. In our research, all E. coli strains showed increased resistance to Amoxicillin (71.4%), Cefotaxime (71.4%), Sulphamethaxzole/Trimethoprim (66.6%), and Tetracycline (52.4%). In a comparative analysis, Yu et al. (2020) showed that every E. coli isolate exhibited resistance to Penicillin and Acetylspiramycin (100%), Lincomycin and Oxacillin (98.8%), and sulphamethaxzole (53%), demonstrating multiple resistance to at least three classes of antimicrobials. Additionally, E. coli strains derived from bovine mastitis showed resistance to Oxytetracycline, Streptomycin, Amoxicillin, Procaine Penicillin, Ampicillin, Tetracycline, Amoxicillin–Clavulanic acid, and Sulfamethoxazole–Trimethoprim (Younis et al., 2017; Ameen et al., 2019; Ismail and Abutarbush, 2020; Tahar et al., 2020). The resistance patterns of E. coli strains that showed multidrug resistance. Among 10 isolates of E. coli strains, 8 (80%) demonstrated MDR against 6 distinct classes of antimicrobial antibiotics, including β-lactams, Aminoglycosides, Macrolides, Tetracyclines, Quinolones, Sulfonamides, and Phenicols. The examined isolates exhibited 6 phenotypes that demonstrated the distribution and relationship between the occurrence of phenotypic patterns and antibiotic resistance genes in E. coli strains sourced from clinical and subclinical mastitis. This finding aligns with Ahmed and Shimamoto (2011), who reported that coliform bacteria from bovine mastitis exhibited MDR in Egypt, including Ent. cloacae (7.1%), K. pneumoniae (6.3%), K. oxytoca (6.3%), E. coli (4.5%), and Cit. freundii (2.7%). In addition, these results align with those of two recent studies in Algeria performed by Tahar et al. (2020) and Ghallache et al. (2021), which indicated multi-drug resistance in the majority of E. coli isolates from milk affected by bovine mastitis. In Indonesia, 7.26% of E. coli isolates from 150 tested milk samples exhibited an MDR profile (Widodo et al., 2022). In Canada, 62.8% of resistant E. coli isolates were MDR. Resistance to >5 antimicrobials was most common among E. coli isolates (Saini et al., 2012). In Bangladesh, 84.2% (16/19) of the E. coli isolates were identified as MDR, particularly Amoxicillin (94.5%), Ampicillin (89.5%), and tetracycline (89.5%). Moreover, the resistance genes tetA and blaTEM-1 were found in 100% and 38.9% of E. coli isolates, respectively (Bag et al., 2021). Four mechanisms of intrinsic resistance are drug uptake, drug target modification, inactivating drug activity, and active drug efflux. These intrinsic changes include the expression of antibiotic-inactivating enzymes, efflux pumps, permeability, and target modifications (Smith et al., 2024). Smith et al. (2024) reported that the E. coli resistance mechanism AcrAB-TolC efflux pump interacts with commonly used antibiotics. A recent study provides important new information on the spread of drug resistance by identifying novel antibiotic-resistant E. coli strains in bovine manure from a Polish dairy farm. Researchers found new plasmids in these bacteria that carry genes resistant to several drugs through whole-genome sequencing. These results are particularly concerning because these plasmids have the ability to propagate among several bacterial species, which could accelerate the development of antibiotic resistance (Zalewska et al., 2024). Adverse outcomes, in field situations, regarding the effectiveness of mastitis treatment are primarily linked to the rise and evolution of antibiotic resistance patterns in livestock (Seyda et al., 2014). As a result, this research explored various antibiotic resistance genes in E. coli isolates and noted that all examined E. coli isolates exhibited 100% resistance to the blaTEM gene. Moreover, the genotypes responsible for Streptomycin (Aada2), tetA, and sul1 resistance showed incidence rates of 50%, 40%, and 50%, respectively, with no presence of the qurA gene detected. In Egypt, Younis et al. (2017) used PCR to identify blaTEM, blaSHV, and blaCTX, with prevalence rates of 100%, 80%, and 60%, respectively, in gram-negative isolates, whereas sul1 was identified in 80% of the isolates tested. In Algeria, Tahar et al. (2020) demonstrated that tetA and blaTEM-1 genes were prevalent at rates of 44.2% and 30.7%, respectively, in E. coli strains collected from dairy farms. Memon et al. (2016) discovered the antibiotic-resistant genes TraT, FimH, papC, iucD, F4 (K88), and sea in 103 E. coli isolates from 22 dairy farms in China, which exhibited a high occurrence of resistance genes CTX-M, qnrS, tetA, tetB, sul1, and sul2. In China, Xu et al. (2023) utilized PCR to screen 12 resistance genes: blaTEM, blaCTX-M, blaSHV, armA, armB, tetA, tetB, tetC, qnrS, qepA, oqxA, and oqxB. All E. coli isolates notably contained the blaTEM gene (100%) along with blaCTX-M (53.8%), armA (9.0%), tetA (26.9%), tetB (2.6%), tetC (20.5%), blaSHV (20.5%), qnrS (29.5%), oqxA (37.2%), and oqxB (1.3%). In Iran, the eaeA and stx1 genes were identified most frequently in 89.3% and 72.3% isolates from mastitic milk samples, respectively (Aflakian et al., 2022). ConclusionIn this study, the prevalence of clinical and subclinical mastitis was 16% and 9%, respectively. Among the isolated coliform bacteria from the examined mastitic cows, E. coli was the most common one. The antibiogram analysis indicated that all isolated E. coli strains showed increased resistance to Amoxicillin, Cefotaxime, Sulphamethoxazole/Trimethoprim, and Tetracycline. A strong susceptibility was observed to Ciprofloxacin, Amoxicillin+Clavulanic, Streptomycin, Gentamicin, Chloramphenicol, and Colistin. The blaTEM gene is among the most prevalent antibiotic resistance genes detected in E. coli isolates (100%). In addition, the genotypes responsible for resistance to tetA, aadA, and sulI were found at rates of 50%, 40%, and 50%, respectively. AcknowledgmentsNone. Conflict of interestNone of the authors has any financial or personal relationships that could inappropriately influence or bias the content of this paper. FundingThis work did not receive any funding from any agency. Author contributionsAll authors contributed equally to the planning of the work, study design, measurement of parameters, and writing of the manuscript. All authors have read, reviewed, and approved the final manuscript. Data availabilityThe article includes all data supporting the study’s findings. In case additional data are required, the corresponding author will provide it upon reasonable request. ReferencesAbebe, R., Hatiya, H., Abera, M., Megersa, B. and Asmare, K. 2016. Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet. Res. 12, 1–11; doi: 10.1186/s12917-016-0905-3 Abed, A.H., Menshawy, A.M.S., Zeinhom, M.M.A., Hossain, D., Khalifa, E., Wareth, G. and Awad, M.F. 2021. Subclinical mastitis in selected bovine dairy herds in north upper egypt: assessment of prevalence, causative bacterial pathogens, antimicrobial resistance and virulence-associated genes. Microorganisms 9 (6), 1175; doi: 10.3390/microorganisms9061175 Abegewi, U.A., Esemu, S.N., Ndip, R.N. and Ndip, L.M. 2022. Prevalence and risk factors of coliform-associated mastitis and antibiotic resistance of coliforms from lactating dairy cows in North West Cameroon. PLoS One. 17, 1–18; doi: 10.1371/journal.pone.0268247 Adzitey, F., Setsoafia, C.K. and Teye, G.A. 2015. Antibiotic susceptibility of Escherichia coli isolated from milk and hands of milkers in Nyankpala community of Ghana. Curr. Res. Dairy Sci. 8(8), 6–11.;doi: 10.3923/crds.2016.6.11 Aflakian, F., Rad, M., Salimizand, H., Nemati, A. and Zomorodi, A.R. 2022. Detection of virulence genes and determination of the antimicrobial susceptibility of Escherichia coli isolates with mastitis in Mashhad, Iran. A short communication Vet. Arhiv. 92(4), 525–530; doi: 10.24099/vet. arhiv.1346 Ahmed, A.M. and Shimamoto, T. 2011. Molecular characterization of antimicrobial resistance in Gram-negative bacteria isolated from bovine mastitis in Egypt. Microbiol. Immunol. 55, 318–327; doi: 10.1111/j.1348-0421.2011.00323.x Ahmed, H.F., Straubinger, R.K., Hegazy, Y.M. and Ibrahim, S. 2018. Subclinical mastitis in dairy cattle and buffaloes among small holders in Egypt: prevalence and evidence of virulence of Escherichia coli causative agent. Trop. Biomed. 35(2), 321–329. Ameen, F., Reda, S.A., El-Shatoury, S.A., Riad, E.M., Enany, M.E. and Alarfaj, A.A. 2019. Prevalence of antibiotic resistant mastitis pathogens in dairy cows in Egypt and potential biological control agents produced from plant endophytic actinobacteria. Saudi J. Biol Sci. 26, 1492–1498; doi: 10.1016/j.sjbs.2019.09.008 Bag, M.A.S., Khan, M.S.R., Sami, M.D.H., Begum, F., Islam, M.S., Rahman, M.M., Rahman, M.T. and Hassan, J. 2021. Virulence determinants and antimicrobial resistance of E. coli isolated from bovine clinical mastitis in some selected dairy farms of Bangladesh. Saudi J. Biol. Sci. 28, 6317–6323; doi: 10.1016/j.sjbs.2021.06.099 Balamurugan, S. and Ranjith, R. 2018. Cold metal transfer ( CMT ) technology—a review. Int. J. Pure Appl. Math. 119, 2185–2196. Bari, M.S., Rahman, M.M., Persson, Y., Derks, M., Sayeed, M.A., Hossain, D., Singha, S., Hoque, M.A., Sivaraman, S., Fernando, P., Ahmad, I., Samad, A. and Koop, G. 2022. Subclinical mastitis in dairy cows in south-Asian countries: a review of risk factors and etiology to prioritize control measures. Vet. Res. Commun. 46(3), 621–640; doi: 10.1007/s11259-022-09948-x Bartlett, P.C., Agger, J.F., Houe, H. and Lawson, L.G. 2001. Incidence of clinical mastitis in Danish dairy cattle and screening for non-reporting in a passively collected national surveillance system. Prev. Vet. Med. 48(2), 73–83; doi: 10.1016/S0167-5877(00)00192-6 Busanello, M., Rossi, R.S., Cassoli, L.D., Pantoja, J.C.F. and Machado, P.F. 2017. Estimation of prevalence and incidence of subclinical mastitis in a large population of Brazilian dairy herds. J. Dairy Sci. 100(8), 6545–6553; doi: 10.3168/jds.2016-12042 Cabral, J.F., Marco, A., ocirc nio, P. da S., Thiago, S.C., Rafaella, B.B., Julliano, C.G., Neves, R.B., Nicolau, E.S. and Lage, M.E. 2015. Procedure for collecting milk sample and the number of milkings in relation to chemical composition and somatic cells of the fresh milk. Afri. J. Agr. Res. 10(50), 4623–4631; doi: 10.5897/AJAR2015.9183 Cheng, W.N. and Han, S.G. 2020. Bovine mastitis: risk factors, therapeutic strategies, and alternative treatments—a review. Asian-Australas J. Anim. Sci. 33(11), 1699–1713; doi: 10.5713/ajas.20.0156 Cowan, D.A., Ramond, J.B., Makhalanyane, T.P. and De Maayer, P. 2015. Metagenomics of extreme environments. Curr. Opin. Microbiol. 25, 97–102; doi: 10.1016/j.mib.2015.05.005 CLSI 2018. Clinical and Laboratory Standards Institute, Wayne. Wayne, PA: CLSI. Available via https://clsi.org/standards/products/microbiology/documents/m11/<AQ3> de Souza, M.M.S., Dubenczuk, F.C., Melo, D.A., Holmström, T.C.N., Mendes, M.B., Reinoso, E.B., Coelho, S.M.O. and Coelho, I.S. 2024. Antimicrobial therapy approaches in the mastitis control driven by one health insights. Braz. J. Vet. Med. 46, e002624; doi: 10.29374/2527-2179.bjvm002624 El-Mohandes, S.S., Eid, R.H., Allam, A.M., Abou-Zeina, H.A.A. and Elbayoumy, M.K. 2022. Phenotyping and genotyping studies on extended-spectrum β-lactamase-producing Escherichia coli isolates from mastitic cows on dairy farms in Egypt. Vet. World. 15, 890–897; doi: 10.14202/vetworld.2022.890-897 Elbably, M. and Asmaa, H. 2013. Risk factors associated with mastitis occurrence in dairy herds in Benisuef, Egypt. World’s Vet. J. 3(1), 5–10; doi: 10.5455/wvj.20130223 ElFaramaway, R., Abdeen, E., Tawab, A. and Mousa, W. 2019. Antibiogram profile and molecular characterization of coa and spa genes of methicillin resistant Staphylococcus aureus (MRSA) from clinical mastitis. Alex. J. Vet. Sci. 61(1), 32–38; doi: 10.5455/ajvs.41507 Endale, H., Mathewos, M. and Abdeta, D. 2023. Potential causes of spread of antimicrobial resistance and preventive measures in one health perspective-a review. Infect. Drug Resist. 16, 7515–7545; doi: 10.2147/IDR.S428837 Food and Drug Administration. 2011. Process validation: general principles and practices. Silver Spring, MD: Food and Drug Administration. Available via https://www.fda.gov/files/drugs/published/Process-Validation--General-Principles-and-Practices.pdf Ghallache, L., Mohamed-Cherif, A., China, B., Mebkhout, F., Boilattabi, N., Bouchemal, A., Ahmed Rebia, Ayachi1, A., Khelef, D., Miroud, K. and Ait-Oudhia, K. 2021. Antibiotic resistance profile of Escherichia coli isolated from bovine subclinical mastitis of dairy farms in Algeria from 2017 to 2019. World’s Vet. J. 11(3), 402–415; doi: 10.54203/scil.2021.wvj52 Goulart, D.B. and Mellata, M. 2022. Escherichia coli mastitis in dairy cattle: etiology, diagnosis, and treatment challenges. Front. Microbiol. 13, 1–15; doi: 10.3389/fmicb.2022.928346 Hussein, O.H., Abdel Hameed, K.G. and El-Malt, L.M. 2022. Prevalence and public health hazards of subclinical mastitis in dairy cows. SVU-Int. J. Vet. Sci. 5(3), 52–64; doi: 10.21608/svu.2022.131652.1189 Ismail, Z.B. and Abutarbush, S.M. 2020. Molecular characterization of antimicrobial resistance and virulence genes of Escherichia coli isolates from bovine mastitis. Vet. World. 13 (8), 1588–1593; doi: 10.14202/vetworld.2020.1588-1593 Kateete, D.P., Kabugo, U., Baluku, H., Nyakarahuka, L., Kyobe, S., Okee, M., Najjuka, C.F. and Joloba, M.L. 2013. Prevalence and antimicrobial susceptibility patterns of bacteria from milkmen and cows with clinical mastitis in and around Kampala, Uganda. PLoS One. 8(5), e63413; doi: 10.1371/journal.pone.0063413 Khasapane, N.G., Byaruhanga, C., Thekisoe, O., Nkhebenyane, S.J. and Khumalo, Z.T.H. 2023. Prevalence of subclinical mastitis, its associated bacterial isolates and risk factors among cattle in Africa: a systematic review and meta-analysis. BMC Vet. Res. 19(1), 123; doi: 10.1186/s12917-023-03673-6 Memon, J., Kashif, J., Hussain, N., Yaqoob, M., Ali, A., Buriro, R., Soomro, J., Hassan, M.F., Sahito, B. and Hongjie, F. 2016. Serotypes, genotypes, virulence factors and antimicrobial resistance genes of Escherichia coli isolated in bovine clinical mastitis from eastern China. Pak. Vet. J. 36(4), 493–498. Mousa, S.W., Zaghawa, A.A., Nayel, M.A., Abdeen, E.E., Elsify, A.M. and Salama, A.A. 2015. Studies on clinical and subclinical mastitis in Menoufia Governate with application of PCR for diagnosis. Minufiya Vet. J. 9, 78–84. Oliveira, C.S.F., Hogeveen, H., Botelho, A.M., Maia, P.V., Coelho, S.G. and Haddad, J.P.A. 2015. Cow-specific risk factors for clinical mastitis in Brazilian dairy cattle. Prev. Vet. Med. 121(3-4), 297–305; doi: 10.1016/j.prevetmed.2015.08.001 Pascu, C., Herman, V., Iancu, I. and Costinar, L. 2022. Etiology of mastitis and antimicrobial resistance in dairy cattle farms in the Western part of Romania. Antibiotics 11(1), 57; doi: 10.3390/antibiotics11010057 Saini, V., McClure, J.T., Léger, D., Keefe, G.P., Scholl, D.T., Morck, D.W. and Barkema, H.W. 2012. Antimicrobial resistance profiles of common mastitis pathogens on Canadian dairy farms. J. Dairy Sci. 95, 4319–4332; doi: 10.3168/jds.2012-5373 Seyda, C., Gökçen, D. and Ünlü, S.M. 2014. Detection of several virulence properties, antibiotic resistance and phylogenetic relationship in E. coli isolates originated from cow mastitis Acta Vet. 64, 413–425; doi: 10.2478/acve-2014-0039 Smith, B.L., Fernando, S. and King, M.D. 2024. Escherichia coli resistance mechanism AcrAB-TolC efflux pump interactions with commonly used antibiotics: a molecular dynamics study. Sci. Rep. 14, 2742; doi: 10.1038/s41598-024-52536-z Stanek, P., Żółkiewski, P. and Januś, E. 2024. A review on mastitis in dairy cows research: current status and future perspectives. Agriculture 14(8), 1292; doi: 10.3390/agriculture14081292 Tahar, S., Nabil, M.M., Safia, T., Ngaiganam, E.P., Omar, A., Hafidha, C., Hanane, Z., Rolain, J. and Diene, S.M. 2020. Molecular characterization of multidrug-resistant Escherichia coli isolated from milk of dairy cows with clinical mastitis in Algeria. J. Food Prot. 83, 2173–2178; doi: 10.4315/JFP-20-198 Tommasoni, C., Fiore, E., Lisuzzo, A. and Gianesella, M. 2023. Mastitis in dairy cattle: on-farm diagnostics and future perspectives. Animals (Basel) 13(15), 2538; doi: 10.3390/ani13152538 Widodo, A., Lamid, M., Effendi, M.H., Khailrullah, A.R., Riwu, K.H.P. and Yustinasari, L.R. 2022. Antibiotic sensitivity profile of multidrug-resistant (MDR) Escherichia coli isolated from dairy cow’s milk in Probolinggo, Indonesia. Biodiversitas 23(10), 4971–4976. Available via https://smujo.id/biodiv/article/view/11896 Xu, T., Cao, W., Huang, Y., Zhao, J., Wu, X. and Yang, Z. 2023. The prevalence of Escherichia coli derived from bovine clinical mastitis and distribution of resistance to antimicrobials in part of Jiangsu Province, China. Agriculture. 13(1), 90; doi: 10.3390/agriculture13010090 Younis, G., Awad, A. and Ashraf, N. 2017. Molecular and phenotypic characterization of antimicrobial resistance in gram negative bacteria recovered from subclinical mastitis. Adv. Anim. Vet. Sci. 5(5), 196–204; doi: 10.17582/journal.aavs/2017/5.5.196.204 Yu, Z.N., Wang, J., Ho, H., Wang, Y.T., Huang, S.N. and Han, R.W. 2020. Prevalence and antimicrobial-resistance phenotypes and genotypes of Escherichia coli isolated from raw milk samples from mastitis cases in four regions of China. J. Global Antimicr. Resist. 22, 94–101; doi: 10.1016/j.jgar.2019.12.016 Zalewska, M., Błażejewska, A., Gawor, J., Adamska, D., Goryca, K., Szeląg, M., Kalinowski, P. and Popowska, M. 2024. A newly identified IncY plasmid from multi-drug-resistant Escherichia coli isolated from dairy cattle feces in Poland. Microbiol. Spectr. 12(8), e0087724; doi: 10.1128/spectrum.00877-24 | ||
| How to Cite this Article |
| Pubmed Style Elsobky Y, Rabah IM, Mousa WS, Sabbah K, Nayel MA, Elsify AM, Elgendy AA, Zaghawa AA, Salam AA, Abu-seida AM, Abdulkarim A, Elkamshishi MM. Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt. Open Vet. J.. 2025; 15(5): 2094-2102. doi:10.5455/OVJ.2025.v15.i5.27 Web Style Elsobky Y, Rabah IM, Mousa WS, Sabbah K, Nayel MA, Elsify AM, Elgendy AA, Zaghawa AA, Salam AA, Abu-seida AM, Abdulkarim A, Elkamshishi MM. Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt. https://www.openveterinaryjournal.com/?mno=239986 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i5.27 AMA (American Medical Association) Style Elsobky Y, Rabah IM, Mousa WS, Sabbah K, Nayel MA, Elsify AM, Elgendy AA, Zaghawa AA, Salam AA, Abu-seida AM, Abdulkarim A, Elkamshishi MM. Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt. Open Vet. J.. 2025; 15(5): 2094-2102. doi:10.5455/OVJ.2025.v15.i5.27 Vancouver/ICMJE Style Elsobky Y, Rabah IM, Mousa WS, Sabbah K, Nayel MA, Elsify AM, Elgendy AA, Zaghawa AA, Salam AA, Abu-seida AM, Abdulkarim A, Elkamshishi MM. Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt. Open Vet. J.. (2025), [cited January 25, 2026]; 15(5): 2094-2102. doi:10.5455/OVJ.2025.v15.i5.27 Harvard Style Elsobky, Y., Rabah, . I. M., Mousa, . W. S., Sabbah, . K., Nayel, . M. A., Elsify, . A. M., Elgendy, . A. A., Zaghawa, . A. A., Salam, . A. A., Abu-seida, . A. M., Abdulkarim, . A. & Elkamshishi, . M. M. (2025) Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt. Open Vet. J., 15 (5), 2094-2102. doi:10.5455/OVJ.2025.v15.i5.27 Turabian Style Elsobky, Yumna, Ibrahim M. Rabah, Walid S. Mousa, Khaled Sabbah, Mohamed A. Nayel, Ahmed M. Elsify, Asmaa A. Elgendy, Ahmed A. Zaghawa, Akram A. Salam, Ashraf M. Abu-seida, Abdulrahman Abdulkarim, and Mohamed M. Elkamshishi. 2025. Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt. Open Veterinary Journal, 15 (5), 2094-2102. doi:10.5455/OVJ.2025.v15.i5.27 Chicago Style Elsobky, Yumna, Ibrahim M. Rabah, Walid S. Mousa, Khaled Sabbah, Mohamed A. Nayel, Ahmed M. Elsify, Asmaa A. Elgendy, Ahmed A. Zaghawa, Akram A. Salam, Ashraf M. Abu-seida, Abdulrahman Abdulkarim, and Mohamed M. Elkamshishi. "Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt." Open Veterinary Journal 15 (2025), 2094-2102. doi:10.5455/OVJ.2025.v15.i5.27 MLA (The Modern Language Association) Style Elsobky, Yumna, Ibrahim M. Rabah, Walid S. Mousa, Khaled Sabbah, Mohamed A. Nayel, Ahmed M. Elsify, Asmaa A. Elgendy, Ahmed A. Zaghawa, Akram A. Salam, Ashraf M. Abu-seida, Abdulrahman Abdulkarim, and Mohamed M. Elkamshishi. "Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt." Open Veterinary Journal 15.5 (2025), 2094-2102. Print. doi:10.5455/OVJ.2025.v15.i5.27 APA (American Psychological Association) Style Elsobky, Y., Rabah, . I. M., Mousa, . W. S., Sabbah, . K., Nayel, . M. A., Elsify, . A. M., Elgendy, . A. A., Zaghawa, . A. A., Salam, . A. A., Abu-seida, . A. M., Abdulkarim, . A. & Elkamshishi, . M. M. (2025) Isolation and molecular identification of multidrug-resistant Escherichia coli strains isolated from mastitic cows in Egypt. Open Veterinary Journal, 15 (5), 2094-2102. doi:10.5455/OVJ.2025.v15.i5.27 |