| Research Article | ||
Open Vet. J.. 2025; 15(4): 1757-1764 Open Veterinary Journal, (2025), Vol. 15(4): 1757-1764 Research Article Isolation and molecular detection of Cyclospora from water samples in Mosul citySenaa Abdullah Ali Al-jarjary1*, Manal H. Hasan2 and Omar Hashim Sheet31Department of Biology, College of Science, University of Mosul, Mosul, Iraq 2Department of Microbiology, College of Veterinary Medicine, University of Mosul, Mosul, Iraq 3Department of Veterinary Public Health, College of Veterinary Medicine, University of Mosul, Mosul, Iraq *Corresponding Author: Senaa Abdullah Ali Al-jarjary. Department of Biology, College of Science, University of Mosul, Iraq. Email: sensbio23 [at] uomosul.edu.iq Submitted: 18/1/2025 Accepted: 29/3/2025 Published: XX/XX/2025 © 2025 Open Veterinary Journal
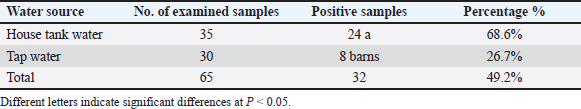
AbstractBackground: Cyclospora can be transferred via water- and food-borne routes, and it causes diseases; therefore, it is considered a major public health concern worldwide. Aim: The objectives of this study were to detect Cyclospora in water using microscopic examination and molecular biology and to determine the relationship between Cyclospora in the current research and Cyclospora registered in GenBank based on the phylogenetic tree. Methods: Sixty-five water samples (35 house tanks and 30 tap water) were collected from Mosul city’s left and right sides between September 2023 and March 2024. Results: The present study indicates that the prevalence of Cyclospora in water in Mosul City, based on microscopic examination, was 49.2%. The highest occurrence of Cyclospora in water was observed on the right side at 54.5%, while the lowest occurrence, at 43.8%, was observed on the left side. Furthermore, the occurrence of Cyclospora was 68.6% in house tank water samples was 68.6%, while in tap water samples, it was 26.7%. Furthermore, the PCR as revealed that Cyclospora was detected in 3.13% (1 of 32) of the water samples collected from Mosul. A novel strain of Cyclospora has been registered in the NCBI GenBank. Conclusion: Proper hygiene during water sterilization and storage can help keep water uncontaminated for an extended period. Keywords: Cyclospora, Water, Molecular detection, Phylogenetic analysis IntroductionCyclospora spp. is considered to be one of the most important waterborne diseases that affect human life in both developed and worldwide (Efstratiou et al., 2017). The protozoan parasites Cyclospora spp. fall under the family Eimeriidae, order Eimeriorina, class Sporozoasida, subclass Coccidiasina, and phylum Apicomplexa. The only confirmed member of the genus Cyclospora that infects citizens is Cyclospora cayetanensis. In the past, organisms linked to human diarrhea were described as having bodies that resembled those of Cyanobacterium or Coccidian, blue–green algae, large Cryptosporidium, or tiny Isospora-like organisms. However, upon closer inspection, these were probably C. cayetanensis (Ortega and Sanchez, 2010). The primary identification and naming of C. cayetanensis occurred 25 years ago (Ortega et al., 1993). In previous years, organisms associated with human diarrhea have been discovered as having blue–green algae, giant Cryptosporidium, microscopic Isospora-like organisms, and Cyanobacterium-like bodies or coccidian-like bodies. Nevertheless, upon closer inspection, these were most likely C. cayetanensis (Ortega and Sanchez, 2010). Several other Cyclospora species have been discovered in nonhuman primates, showing that they are likely host-specific (Eberhard et al., 1999). First reported three cases of Cyclosporiasis in patients in Basra, Iraq in 1999 (Ali and Mahdi, 1999). The most recently discovered species is Cyclospora macacae, which has been identified in rhesus monkeys (Macaca mulatta) (Li et al., 2015). Based on ITS-2 analysis, C. oocysts have been identified in chimpanzees (Pan troglodytes) and Cynomolgus monkeys (Macaca fascicularis), and the sequences of them matched those found in C. cayetanensis. When determining the species status of these parasites, attention must be given because there remain uncertainties about their host specificity and capacity for human transmission (Marangi et al., 2015). Animal excrement has been discovered to include Oocysts that resemble Cyclospora in a variety of species, including chickens, dogs, rats, monkeys, mice, ducks, and other avian species (Cordãn et al., 2008; Cordãn et al., 2009). Furthermore, based on molecular data, Oocysts of Cyclospora spp. have also been discovered in dairy cattle manure (Li et al., 2007). Although the main mode of transmission of Cyclospora is fecal–oral, other processes of transmission are still not fully understood (Ortega et al., 1998). Direct transfer from one person to another is improbable. Indirect transmission may occur when an infected individual contaminates the surroundings; the Oocysts sporulate when appropriate circumstances are met, and contaminated food or drinking water is subsequently consumed. It has also been suggested that soil plays a role in transmission (Chacín-Bonilla, 2008). Cyclospora, Microsporidia, and Cryptosporidium have been discovered in drinking water, effluent, and recreational water, according to several investigations (Dowd John et al., 2003; Tekeli et al., 2024; Al-Jarjary et al., 2024). In addition, they are associated with global waterborne outbreaks (Rabold et al., 1994). The U.S. The Environmental Protection Agency’s drinking water contaminant candidate lists, contaminant candidate list 2 )CCL2( and CCL3, contain both Cyclospora and Microsporidia (Enterocytozoon bieneusi, Encephalitozoon intestinalis, Encephalitozoon cuniculi, Encephalitozoon hellem, and Vittaforma corneae) because of the possibility for water borne communication (US Environmental Protection Agency, 2005; US Environmental Protection Agency, 2009). Cryptosporidium spp. are significant parasitic protozoa related to outbreaks of waterborne diseases in Europe and the United States (Feng and Xiao, 2011). The potential of Cyclospra transmission through water contaminated via human sewage or wastewater was discovered by environmental surveillance studies performed in the 1990s, which reported the detection of C. cayetanensis Oocysts in wastewater in Peru and in rivers impacted by raw sewage outfalls in Guatemala (Sturbaum et al., 1998). Numerous disease outbreaks attributed to Cyclospora have been discovered in earlier studies (Baldursson and Karanis, 2011). Cyclosporiasis infections may appear asymptomatic or symptomatic, with the majority of patients who are symptomatic exhibiting acute, self-limiting symptoms. Cyclosporiasis is often accompanied by cramps, nausea, diarrhea, and weight loss. If left untreated, clinical symptoms may persist for a few days, a month, or more. Additionally, those with weakened immune systems may constantly exhibit symptoms (Pape et al., 1994). Factors associated with risk like age, underlying health conditions that affect immunity, prior exposure, and financial status, can affect the presence and severity of symptoms (Almeria et al., 2019). Numerous techniques are available to recognize Cyclospora oocysts based on their shape and autofluorescence characteristics (Dixon et al., 2005). Modified Ziehl–Neelsen acid-fast staining is a frequently employed microscopic inspection stain and is recommended for the identification of Cyclospora Oocysts (Brennan et al., 1996). Cyclospora serological tests for screening might be beneficial in identifying outbreaks as well as assisting epidemiological research (Ortega and Sanchez, 2010). Nevertheless, at present, no serological tests are accessible to assess human exposure to Cyclospora. Molecular biological analyses are among the most well-known methods based on parasite DNA sequences; they are thought to be more precise and less straightforward than other methods. Based on SSU rRNA gene sequences, (Reiman et al., 1996) created the first PCR technique employed for the clinical determination of C. cayetanensis. Subsequently, numerous more PCR assays have been created. With specificity, SSUU rRNA gene-based real-time PCR can recognize DNA from as few as oocysts (Varma et al., 2003). This study’s primary objectives were to recognize Cyclospora spp. in water samples using microscopic and molecular techniques, to determine the relationships between Cyclospora using region-specific sequencing analysis, and to determine the genetic diversity of Cyclospora found in additional sources. Materials and MethodsSamplingAll water samples employed in this investigation have been collected between September 2023 and March 2024 from multiple locations in Mosul city, Iraq. 65 water samples (35 house tanks and 30 tap water) have been obtained from Mosul city’s left and right sides during the study period. The left side of the city included Almagmoaa, Hay Alsuker, Hay Alarabe, Hay Albaladeat, and Hay Almuthana, while the right side included Hay Althora, Mosul Algededa, Aldwasa, Hay Alamel, and Hay Alshefaa. Each water sample had a volume of 250–500 ml. Each sample was collected using sterilized containers. The samples were sent to the central laboratory of the College of Veterinary Medicine, University of Mosul for examination. Microscopic examinationThe water samples were examined by direct wet smearing, analysis of Lugol’s iodine, and sedimentation. All slides were prepared and examined under a light microscope at different magnifications (10x, 40x, and 100x) to detect the parasite Cyclospora spp. (Bakir et al., 2003; John and Petri, 2006). The modified Ziehle-Nelseen stain was used to detect Oocysts of Cyclospora spp. according to Baron et al. (1994). Statistical analysisThe prevalence of parasites in the water samples was assessed, and the chi-square (χ2) test was employed to evaluate statistical significance at a threshold of P < 0.05. All data were analyzed using SPSS (Snedecor and Cochran, 1989). Water sample preparation for PCR ExaminationThe container (150 ml) was filled with 100 ml of water and tested positive using a microscope. The supernatant and sediment sections were separated using a refrigerated centrifuge running at high speed for 20 min. The supernatant was then disregarded to remove any residual sediment. This procedure was repeated several times to purify the parasite, and the sediment was placed in sterile, clean tubes and kept at –20°C for the PCR test. DNA extractionAs previously described (Li et al., 2007), with a few slight modifications, the DNA extracts were examined for the presence of Cyclospora spp. using nested PCR amplification of a 500 bp fragment of the small subunit ribosomal RNA (SSUrRNA) gene. The primer forward (5′-AATGTAAAACCCTTCCAGAGTAAC-3′) and reverse : (5′ GCAATAATCTATCCCCATCAC G-3′) primers were used in the first cycle of PCR, with the following cycling variables: a 7-min hot start at 94°C, 35 cycles of 95°C for 45 s, 55°C for 45 s, 72°C for 90 s, and a final extension at 72°C for 10 min. An internal primer forward (5′-AATTCCAGCTCCAATAGTGTAT-3′) and reverse (5′-CAGGAGAAGCCAAGGTAGGCRTTT-3′) were used for the second cycle of PCR. Except for the 1-min extension period, the thermal cycling conditions were the same as those of the first-round PCR. The entire reaction volume used for PCR amplification was 25 μL. 12.5 μl of 2× GoTaq (Green Mix Master) from Addbio (Korea), 1 μl each of primer F and R, 8.5 μl of double distillate water from Addbio (Korea), and 2 μl of the Cyclospora DNA template were included in the reaction mixture. Gel electrophoresis was then used to visualize the target sequence amplicons. Gel electrophoresis was performed using 2% agarose gels produced by Addbio (Korea), and a 100 bp ladder DNA marker (Addbio, Korea) was added to the wells containing the DNA samples. Electrophoresis was used to separate and visualize the amplified DNA fragments, and a DNA ladder was used to quantify their sizes. DNA sequencingA single PCR amplicon was obtained from water samples previously confirmed to be positive for Cyclospora spp. using conventional PCR. The DNA amplicon was subsequently sequenced by Macrogen (a commercial sequencing company based in South Korea. Using the Basic Local Alignment Search Tool (BLAST) server, the nucleotide sequences from this study were analyzed and compared for similarity with reference strains available in the GenBank database. The NCBI BLASTn tool (http://www.ncbi.nlm.nih.gov) was used to analyze clospora sequences previously published in GenBank. Further alignment and comparison of these sequences were performed using the online multiple sequence alignment program CLUSTALW (MEGA11). This comprehensive approach aimed to elucidate the genetic relationships between the Cyclospora isolates identified in this study and those identified in previous studies. Through purification, sequencing, and subsequent bioinformatics analyses, this research contributes to a deeper understanding of the evolutionary background of these isolates. Ethical approvalNot needed for this study. ResultsBased on microscopic analysis, the results of the current investigation demonstrated that 49.2% (32/65) of the Oocysts of Cyclospora spp. were identified in the water samples based on the morphology of the Oocysts (Figure 1). The high concentration of Oocysts of Cyclospora spp. found in water samples from various regions on the right side of Mosul city was 54.5% (18/33) whereas the lower concentration of Oocysts of Cyclospora spp. found in water samples from different regions on the left side of Mosul city was 43.8% (14/32) with no significant differences as shown in Table 1. In addition, the higher percentage of Oocysts of Cyclospora spp. found in the water samples of house tank water was 68.6% (24/35) whereas the lower percentage of parasites found in tap water was 26.7% (8/30), with significant differences at P < 0.05, as shown in Table 2.
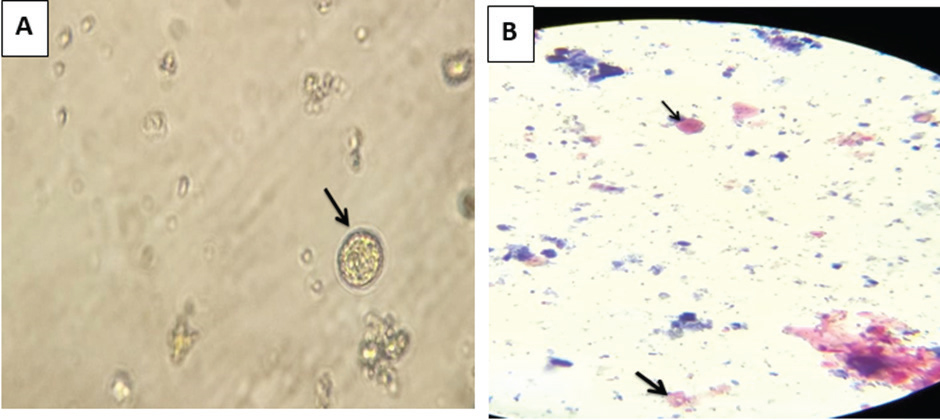
Fig. 1. A: Unstained oocysts of Cyclospora spp. detected from water smears, B: Oocysts of Cyclospora spp. in acid-fast stain. Table 1. Number and percentage of oocyst parasites in water on both sides of Mosul.
Table 2. Number and percentage of oocyst parasites in water samples based on water source
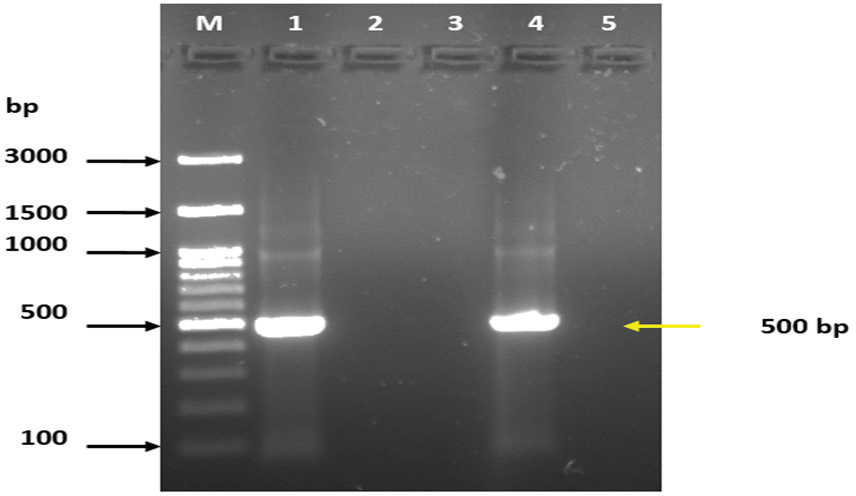
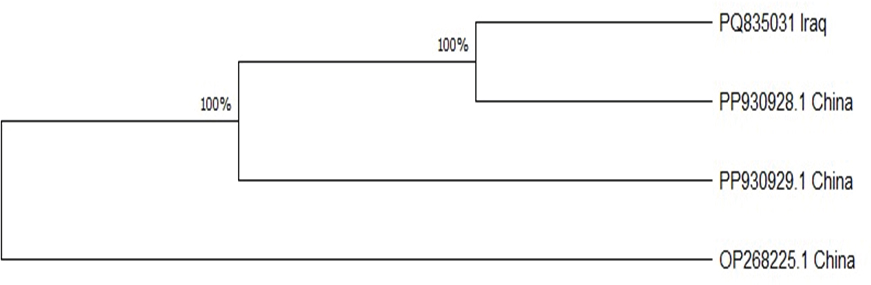
In addition, the results of the PCR assay indicated that the prevalence of Cyclospora spp. in water collected from Mosul was 3.13% (1/32) as shown as Figure 2. In addition, individual sequencing analysis (BLASTn) was performed on a novel gene sequence (SSU rRNA) from Cyclospora spp. isolated from water samples collected in Mosul. The sequencing results of this study correspond to the Cyclospora sequences available in the NCBI GenBank database (accession number PQ835031. According to the MEGA11 program, the relationship between PQ835031 and PP930928, PP930929, and PP268225 from China was found to be strongly supported at 100% (Figure 3). Based on the evolutionary relationships among taxa were inferred using the Neighbor-Joining method (Saitou and Nei, 1987), and the resulting optimal tree is presented. Bootstrap values calculated from 500 replicates are displayed above the branches, indicating the percentage of replicate trees in which the associated taxa clustered together (Felsenstein, 1985). Evolutionary distances were estimated using the Maximum Composite Likelihood method (Tamura et al., 2004) and are expressed as the number of base substitutions per site. The proportion of sites with at least one unambiguous base present in at least one sequence for each descendant clade is indicated next to each internal node in the tree. This analysis included four nucleotide sequences with ambiguous positions removed on a pairwise basis (pairwise deletion option). The final dataset comprised 451 positions. All evolutionary analyses were performed using MEGA11 software (Tamura et al., 2021). DiscussionWater pollution is a major global issue for both human and animal health, as it is directly linked to the prevalence of certain parasitic diseases caused by etiologic agents present in contaminated water. Cyclospora is considered one of the most significant water- and food-borne parasites affecting public health, particularly among young children, the elderly, and individuals who are immunocompromised or immunosuppressed, in both developed and developing countries (Ortega and Sanchez, 2010; Fletcher et al., 2012). The results of this study indicated that the prevalence of Cyclospora based on microscopic test results was 49.2% (32/65), which may be because the water treatment facilities in Mosul city rely on conventional processes, including coagulation, flocculation, sedimentation, filtration, disinfection, and pH adjustment. Several studies in Iraq reported that the prevalence of Cyclospora spp. in drinking water from various cities was 10% in Basra (Al-Mounas et al., 2020), 1.9% in Babylon (Al-Morshidy and Al-Amari, 2015), and 1.38% in Baghdad (Al-Sagur et al., 2015). Previous studies have suggested that the prevalence of Cyclospora spp. in water was lower than that in this study. For instance, in Malaysia, the prevalence of Cyclospora spp. in water was reported to be 11.1% (Bilung et al., 2017), in Vietnam, the prevalence of Cyclospora spp. from water in markets was 11.8 (34/288), and from farms was 8.4% (24/287) (Tram et al., 2008). The high percentage of Cyclospora spp. observed in this study, compared to other studies, may be attributed to unhygienic practices during water cleaning and disinfection, or the reliance on traditional methods (Al-Morshidy and Al-Amari, 2015). The river water contains large amounts of Cyclospora spp., suggesting that it may be contaminated by the feces of humans and animals, including ruminants, dogs, cats, and birds (Lanata, 2003; Plutzer and Karanis, 2016). In addition, the PCR analysis revealed that the prevalence of Cyclospora spp. was 3.13% (1/32). Many recent studies have declared that the prevalence of Cyclospora based on the PCR assay in Italy was 21.3% in treated water and 6.2% in well water (Giangaspero et al., 2015), whereas in Spain, the prevalence of Cyclospora spp. in drinking, river water, and wastewater was 6%, 13%, and 2%, respectively (Galvãn et al., 2013). The development of new detection methods is essential for gaining a better understanding of parasite dispersion in the environment and aiding outbreak investigations (Murphy et al., 2018; Almeria et al., 2025). Several factors have influenced the global prevalence of Cyclospora, including environmental conditions, geographic distribution, hygiene practices, and the educational level of local populations. Additionally, the contamination of household tank water by parasites is significantly affected by the socioeconomic status of the community (Alam et al., 2014; Al-Morshidy and Al-Amari, 2015). The present study utilized microscopic testing and molecular biology techniques to identify Cyclospora in water. Molecular techniques offer greater accuracy and speed, producing results for the identification of microorganisms within three to five hours (Ahmed et al., 2020). Additionally, analyzing DNA sequences of amplicons is a common method used in water sample testing to provide more detailed information about parasites and to enhance detection based on PCR (Durigan et al., 2014; Durigan et al., 2017; Arida et al., 2023). Sanger sequencing will be conducted on any positive samples to verify the presence or absence of Cyclospora spp. in water (Temesgen et al., 2019). Furthermore, the phylogenetic tree revealed a similarity between the Cyclospora spp. identified in this study and those found in China, suggesting the potential for Cyclospora spp. to spread to various regions around the world.
Fig. 2. Agarose gel electrophoresis (2%) showing the typical amplicon of the SSUrRNA gene product of Cyclospora spp. The DNA amplification appeared as a ladder-like pattern. Lane M is the DNA Marker 100 bp ladder (Addbio, Korea), Lane 1 is the positive control, Lanes 2 and 3 represent negative isolates, Lane 4 represents positive isolate, and Lane 5 is a non-template control.
Fig. 3. Analysis of the phylogenetic relationships among Cyclospora spp. in water samples from Mosul, Iraq, using neighbor-joining (NJ) data in MEGA 11. ConclusionThis was an extensive study conducted across a wide range of geographic areas in Mosul, Iraq. Cyclospora spp. were detected in tap and household tank water samples, indicating that water was pumped into homes without proper hygienic measures such as filtration, cleaning, and disinfection. Additionally, the use of unclean tankers or storage in dirty containers can contaminate water with Cyclospora spp. during transmission, which helps to provide suitable conditions for the growth and amplification of Cyclospora spp. and causes various diseases in humans and animals. The phylogenetic tree reveals the spread of Cyclospora spp. across different countries, indicating a relationship between the strains found in these regions. AcknowledgmentThe author extends heartfelt gratitude to Mosul University, particularly the College of Veterinary Medicine and the College of Science, for their generous provision of facilities essential to the completion of this study. Authors’ contributionSenaa Abdullah Ali Al-jarjary, M.H. Hasan and Omar Hashim Sheet. Conceptualization: Senaa Abdullah Ali Al-jarjary and M.H. Hasan: Study design. Senaa Abdullah Ali Al-jarjary collected and analyzed data. Omar Hashim Sheet: M.H. Hasan, and Senaa Abdullah Ali Al-jarjary: Examination samples. Statistical Analysis: M.H. Hasan and Omar Hashim Sheet. Writing: Omar Hashim Sheet, M.H. Hasan and Senaa Abdullah Ali Al-jarjary. Conflict of interestWe will promise no conflicts of interest. FundingThis research received no specific grant from any funding agency. Data availabilityAll relevant data are provided in the manuscript as follows. ReferencesAhmed, I.M., Al-Sanjary, R.A. and Alkazaly, H.H. 2020. Detection of Mycobacterium paratuberculosis in raw cows’ milk using polymerase chain reaction (PCR) technique. Iraqi J. Vet. Sci. 34(1), 83–86.6. DOI:10.33899/ijvs.2019.125556.1075 Alam, M.S., Khan, S.U., Ayaz, S., Khan, M.A., Ahmad, I., Idrees, M. and Waqar, M. 2014. Molecular detection of Giardia lamblia and Cryptosporidium parvum in different water sources of district bantu, Khyber Pakhtunkhwa, Province of Pakistan. Br. Microbiol. Res. J. 4 (1), 80–88. DOI: 10.9734/BMRJ/2014/3947 Ali, N.H. and Mahdi, N.K. 1999. First reported three cases of cyclosporiasis in Iraq. East Mediterr Health J. 5(5), 1055–1057. Al-Jarjary, S.A.A., Hasan, M.H. and Ibrahim, H.B. 2024.Detection of Cryptosporidium spp. and Giardia spp. from water samples in Mosul City, Iraq. Med. J. Babylon. 21(Suppl 2), S282–S287. DOI: 10.4103/MJBL.MJBL_1311_23 Almeria, S., Cinar, H.N. and Dubey, J.P. 2019. Cyclospora cayetanensis and cyclosporiasis: an update. Microorganisms. 7(9), 317. DOI: 10.3390/microorganisms7090317 Almeria, S., Grocholl, J., Mullins, J., Durigan, M., Ewing-Peeples, L., Lauren Rogers, E. Hirneisen, K., Madson, S. and Steven Wang, S. 2025. Multi-laboratory validation of a modified real-time PCR assay (Mit1C) for the detection of Cyclospora cayetanensis in fresh produce. Food Microbiol. 128, 104727. Doi:10.1016/j.fm.2025.104727 Al-Morshidy, K.A. and Al-Amari, M.J. 2015. Detection of parasitic contamination in the drinking water/Babylon province/Iraq. Adv Nat Appl Sci. 1(9), 80–84. Al-Mounas, M.S., Hussein, A.F. and Dawood, N.J. 2020. Distribution of protozoan parasites in drinking water in some regions of Basra Province. Int J Psychosoc Rehabil. 24(04), 3375–3379. DOI:10.37200/IJPR/V24I4/PR201451 Al-Sagur, I.M., Mahmood, S.H. and Al-Obaidi, H.M. 2015. Investigation of Giardia lamblia and other parasites in tap water as a potential source of transmission in some regions of Baghdad. Iraqi J. Sci. 56, 337–344. Arida, J., Shipley, A. and Almeria, S. 2023. Molecular Detection of Cylospora Cyatanensis in two main types of farm soil using real- time PCR assays and method modification for commercial potting mix. Microorganisms. 11(6), 1506. DOI:10.3390/microorganisms11061506 Bakir, B., Tanyuksel, M., Saylam, F., Tanriverdi, S., Engin Araz, R., Hacim, A.K. and Hasde, M. 2003. Investigation of waterborne parasites in drinking water sources in Ankara, Turkey. J. Microbiol. 41(2), 148–151. https://koreascience.kr/article/JAKO200311921740875 Baldursson, S. and Karanis, P. 2011. Waterborne transmission of protozoan parasites: review of worldwide outbreaks–an update 2004–2010. Water Res. 45(20), 6603–6614. DOI: 10.1016/j.watres.2011.10.013 Baron, E.J., Peterson, L.R. and Finegold, S.M. 1994. Diagnostic microbiology. 9th ed. St. Louis, MO: Mosby- Yearbook. Inc. p: 792. https://www.scirp.org/reference/referencespapers? Referenceid=933202 Bilung, L.M., Tahar, A.S., Yunos, N.E., Apun, K., Lim, Y.A., Nillian, E. and Hashim, H.F. 2017. Detection of Cryptosporidium and Cyclospora Oocysts from environmental water for drinking and recreational activities in Sarawak, Malaysia. Biomed Res. Int. 1, 4636420. DOI: 10.1155/2017/4636420 Brennan, M.K., MacPherson, D.W., Palmer, J. and Keystone, J.S. 1996. Cyclosporiasis: a new cause of diarrhea. CMAJ: Can. Med. Assoc. J. 155(9), 1293. https://pubmed.ncbi.nlm.nih.gov/8911296/ Chacín-Bonilla, L. 2008. Transmission of Cyclospora cayetanensis infection: a review focusing on soil-borne cyclosporiasis. Trans. R. Soc. Trop. Med. Hyg. 102(3), 215–216. DOI: 10.1016/j.trstmh.2007.06.005 Cordãn, G.P., Prados, A.H., Romero, D., Moreno, M.S., Pontes, A., Osuna, A. and Rosales, M.J., 2008. Intestinal parasitism in the animals of the zoological garden “Peãa Escrita” (Almuãecar, Spain). Vet. Parasit. 156(3–4), 302–309. DOI: 10.1016/j.vetpar. 2008.05.023 Cordãn, G.P., Prados, A.H., Romero, D., Moreno, M.S., Pontes, A., Osuna, A. and Rosales, M.J. 2009. Intestinal and hematic parasitism in the birds of the Almuãecar (Granada, Spain) ornithological garden. Vet. Parasit. 12, 165(3–4), 361–366. DOI: 10.1016/j.vetpar.2009.07.027 Dixon, B. R., Bussey, J. M., Parrington, L. J. and Parenteau, M. 2005. Detection of Cyclospora cayetanensis Oocysts in human fecal specimens by flow cytometry. J. Clin. Microbiol. 43(5), 2375–2379. DOI: 10.1128/jcm.43.5.2375-2379.2005 Dowd John D., Eliopolus, J., Gerba, C.P., Naranjo, J., Klein, R., Lopez, B., de Mejía, M., Mendoza, C.E. and Pepper, I.L. 2003. Confirmed detection of Cyclospora cayetanensis, Encephalitozoon intestinalis, and Cryptosporidium parvum in water used for drinking. J. Water Health. 1(3), 117–123. DOI:10.2166/wh.2003.0014 Durigan, M., Abreu, A.G., Zucchi, M.I., Franco, R.M. and de Souza, A.P. 2014. Genetic diversity of Giardia duodenalis: multilocus genotyping reveals zoonotic potential between clinical and environmental sources in a metropolitan region of Brazil. PloS one. 9(12), e115489. DOI:10.1371/journal.pone.0115489. Durigan, M., Ciampi-Guillardi, M., Rodrigues, R.C., Greinert-Goulart, J.A., Siqueira-Castro, I.C., Leal, D.A., Yamashiro, S., Bonatti, T.R., Zucchi, M.I., Franco, R.M. and de Souza, A.P. 2017. Population genetic analysis of Giardia duodenalis: genetic diversity and haplotype sharing between clinical and environmental sources. Microbiol. Open. 2:e00424. DOI:10.1002/mbo3.424. Eberhard, M.L., da Silva, A.J., Lilley, B.G. and Pieniazek, N.J. 1999. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci spp. n., C. colobi spp. n., and C. papionis spp. Emerg. Infect. Dis. 5(5), 651. DOI: 10.3201/eid0505.990506 Efstratiou, A., Ongerth, J.E. and Karanis, P. 2017. Waterborne transmission of protozoan parasites: review of worldwide outbreaks-an update 2011–2016. Water Res. 1(114), 14–22. DOI: 10.1016/j.watres.2017.01.036 Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39(4), 783–791. DOI:10.1111/j.1558-5646.1985.tb00420.x Feng, Y. and Xiao, L. 2011. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 24(1), 110–140. DOI: 10.1128/ cmr.00033-10 Fletcher, S.M., Stark, D., Harkness, J. and Ellis, J. 2012. Enteric protozoa in developed countries: A public health perspective. Clin. Microbiol. Rev. 25(3), 420–449. DOI:10.1128/cmr.05038-11 Galvãn, A.L., Magnet, A., Izquierdo, F., Fenoy, S., Rueda, C., Fernandez Vadillo, C., Henriques-Gil, N. and del Aguila, C. 2013. Molecular characterization of human-pathogenic microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: a year-long longitudinal study. Appl. Environ. Microbiol. 79(2), 449–459. DOI:10.1128/AEM.02737-12. Giangaspero, A., Marangi, M., Koehler, A.V., Papini, R., Normanno, G., Lacasella, V., Lonigro, A. and Gasser, R.B. 2015. Molecular detection of Cyclospora in water, soil, vegetables, and humans in southern Italy indicates the need for improved monitoring by health authorities. Int. J. Food Microbiol. 211, 95–100. DOI:10.1016/j.ijfoodmicro.2015.07.002. John, D.T., Petri, W.A., Markell, E. K. and Voge’s, M. 2006. Medical parasitology. 9th edition. pp.107. Available via https://shop.elsevier.com/books/markell-and-voges-medical-parasitology/john/978-0-7216-4793-7 Lanata, C.F. 2003. Studies on food hygiene and diarrheal disease. Int. J. Environ. Health Res. 13(sup1), S175–183. DOI:10.1080/0960312031000102921. Li, G., Xiao, S., Zhou, R., Li, W. and Wadeh, H. 2007. Molecular characterization of Cyclospora-like organism from dairy cattle. Parasitol. Res. 100, 955–961. DOI: 10.1007/s00436-006-0380-z. Li, N., Ye, J., Arrowood, M.J., Ma, J., Wang, L., Xu, H., Feng, Y. and Xiao, L. 2015. Identification and morphological and molecular characterization of Cyclospora macacaen. sp. from rhesus monkeys in China. Parasitol. Res. 114, 1811–1816. DOI: 10.1007/ s00436-015-4367-5 Marangi, M., Koehler, A.V., Zanzani, S.A., Manfredi, M.T., Brianti, E., Giangaspero, A. and Gasser, R.B. 2015. Detection of Cyclospora in captive chimpanzees and macaques using a quantitative PCR-based mutation scanning approach. Paras Vectors. 8, 1–5. DOI 10.1186/s13071-015-0872-8 Murphy, H.R., Cinar, H.N., Gopinath, G., Noe, K.E., Chatman, L.D., Miranda, N.E., Wetherington, J.H., Neal-McKinney, J., Pires, G.S., Sachs, E. and Stanya, K.J. 2018. Interlaboratory validation of an improved method for the detection of Cyclospora cayetanensis in produce using real-time PCR assay. Food Microbiol. 69, 170–178. DOI:10.1016/j.fm.2017.08.008. Ortega, Y.R. and Sanchez, R. 2010. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin. Microbiol. Rev. 23(1), 218–234. DOI: 10.1128/cmr.00026-09 Ortega, Y.R., Sterling, C.R. and Gilman, R.H. 1998. Cyclospora cayetanensis. Adv. Parasitol. 40, 399–418. DOI:10.1016/S0065-308X(08)60128-1 Ortega, Y.R., Sterling, C.R., Gilman, R.H., Cama, V.A. and Diaz, F. 1993. Cyclospora species: a new protozoan pathogen of humans. N. Engl. J. Med. 6, 328(18), 1308–1312. DOI: 10.1056/NEJM199305063281804 Pape, J.W., Verdier, R.I., Boncy, M., Boncy, J. and Johnson, W.D. 1994. Cyclospora infection in adults with HIV infection: clinical manifestations, treatment, and prophylaxis. Ann. Intern. Med. 1, 121(9), 654–657. DOI:10.7326/0003-4819-121-9-199411010-00004 Plutzer, J. and Karanis, P.2016. Neglected waterborne parasitic protozoa and their detection in water. Water Res. 101, 318–332. DOI:10.1016/j.watres.2016.05.085. Rabold, J.G., Hoge, C., Shlim, D., Kefford, C., Rajah, R. and Echeverria, P. 1994. Cyclospora outbreak associated with chlorinated drinking water. Lancet. 344(8933), 1360–1361. DOI:10.1016/S0140-6736(94)90716-1 Reiman, D.A., Schmidt, T.M., Gajadhar, A., Sogin, M., Cross, J., Yoder, K., Sethabutr, O. and Echeverria, P. 1996. Molecular phylogenetic analysis of Cyclospora, a human intestinal pathogen, suggested that it is closely related to Eimeria species. J. Infect. Dis. 173(2), 440–445. DOI:10.1093/infdis/173.2.440 Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4(4), 406–425. Snedecor, G.W. and Cochran, W.G. 1989. Statistical methods. 8th Edition, Ames: Iowa State University Press. Sturbaum, G.D., Ortega, Y.R., Gilman, R.H., Sterling, C.R., Cabrera, L and Klein, D.A. 1998. Detection of Cyclospora cayetanensis in wastewater. Appl. Environ. Microbiol. 64(6), 2284–2286. DOI:10.1128/AEM.64.6.2284-2286.1998 Tamura, K., Nei, M. and Kumar, S. 2004. Prospects for inferring very large phylogenies using the neighbor-joining method. Proc. Natl. Acad. Sci. 101(30), 11030–11035. DOI:10.1073/pnas.0404206101 Tamura, K., Stecher, G. and Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38(7), 3022–3027. DOI:10.1093/molbev/msab120 Tekeli, N.T.F. Al-jarjary, S.A.A., Sheet, O.H. 2024. Molecular detection and phylogenetic analysis of Microsporidia in water and soil in Mosul city. Open Vet. J. Vol. 14(9), 2421–2432. DOI: 10.5455/OVJ.2024.v14.i9.31. Temesgen, T.T., Tysnes, K.R., Robertson, L.J. 2019. A new protocol for the molecular detection of Cyclospora cayetanensis as a contaminant of berry fruits. Front. Microbiol. 10:1939. DOI:10.3389/fmicb.2019.01939. Tram, N.T., Hoang, L.M., Cam, P.D., Chung, P.T., Fyfe, M.W., Isaac-Renton, J.L., Ong, C.S. 2008. Cyclospora spp. in herbs and water samples collected from markets and farms in Hanoi, Vietnam. Trop. Med. Int. Health. 13(11):1415–1420. DOI:10.1111/j.1365-3156.2008.02158.x US Environmental Protection Agency. 2005. Environmental Protection Agency. Contaminant candidate list 2 and regulatory determinations. US Environmental Protection Agency, Washington, DC. http://water.epa.gov/scitech/drinkingwater/dws/ccl/ccl2.cfm. US Environmental Protection Agency. 2009. Environmental Protection Agency. Contaminant candidate list 3. US Environmental Protection Agency, Washington, DC. http://water.epa.gov/scitech/drinkingwater/dws/ ccl/ccl3.cfm. Varma, M., Hester, J.D., Schaefer III, F.W., Ware, M.W. and Lindquist, H.A. 2003. Detection of Cyclospora cayetanensis using quantitative real-time PCR assay. J. Microbiol. Methods. 53(1), 27–36. DOI:10.1016/S0167-7012(02)00209-9. | ||
| How to Cite this Article |
| Pubmed Style Al-jarjary SAA, Hasan MH, Sheet OH. Isolation and molecular detection of Cyclospora from water samples in Mosul city. Open Vet. J.. 2025; 15(4): 1757-1764. doi:10.5455/OVJ.2025.v15.i4.27 Web Style Al-jarjary SAA, Hasan MH, Sheet OH. Isolation and molecular detection of Cyclospora from water samples in Mosul city. https://www.openveterinaryjournal.com/?mno=238413 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i4.27 AMA (American Medical Association) Style Al-jarjary SAA, Hasan MH, Sheet OH. Isolation and molecular detection of Cyclospora from water samples in Mosul city. Open Vet. J.. 2025; 15(4): 1757-1764. doi:10.5455/OVJ.2025.v15.i4.27 Vancouver/ICMJE Style Al-jarjary SAA, Hasan MH, Sheet OH. Isolation and molecular detection of Cyclospora from water samples in Mosul city. Open Vet. J.. (2025), [cited January 25, 2026]; 15(4): 1757-1764. doi:10.5455/OVJ.2025.v15.i4.27 Harvard Style Al-jarjary, S. A. A., Hasan, . M. H. & Sheet, . O. H. (2025) Isolation and molecular detection of Cyclospora from water samples in Mosul city. Open Vet. J., 15 (4), 1757-1764. doi:10.5455/OVJ.2025.v15.i4.27 Turabian Style Al-jarjary, Senaa Abdullah Ali, Manal H. Hasan, and Omar Hashim Sheet. 2025. Isolation and molecular detection of Cyclospora from water samples in Mosul city. Open Veterinary Journal, 15 (4), 1757-1764. doi:10.5455/OVJ.2025.v15.i4.27 Chicago Style Al-jarjary, Senaa Abdullah Ali, Manal H. Hasan, and Omar Hashim Sheet. "Isolation and molecular detection of Cyclospora from water samples in Mosul city." Open Veterinary Journal 15 (2025), 1757-1764. doi:10.5455/OVJ.2025.v15.i4.27 MLA (The Modern Language Association) Style Al-jarjary, Senaa Abdullah Ali, Manal H. Hasan, and Omar Hashim Sheet. "Isolation and molecular detection of Cyclospora from water samples in Mosul city." Open Veterinary Journal 15.4 (2025), 1757-1764. Print. doi:10.5455/OVJ.2025.v15.i4.27 APA (American Psychological Association) Style Al-jarjary, S. A. A., Hasan, . M. H. & Sheet, . O. H. (2025) Isolation and molecular detection of Cyclospora from water samples in Mosul city. Open Veterinary Journal, 15 (4), 1757-1764. doi:10.5455/OVJ.2025.v15.i4.27 |