| Research Article | ||
Open Vet. J.. 2025; 15(8): 3737-3745 Open Veterinary Journal, (2025), Vol. 15(8): 3737-3745 Research Article Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattleDarniati Darniati1*, Ekowati Handharyani2 and Surachmi Setiyaningsih31Laboratory of Microbiology, Faculty of Veterinary Medicine, Syiah Kuala University, Banda Aceh, Indonesia 2Division of Veterinary Clinic Reproduction and Pathology, Faculty of Veterinary Medicine, Institut Pertanian Bogor, Bogor, Indonesia 3Division of Medical Microbiology, School of Veterinary Medicine and Biomedical Sciences, Institut Pertanian Bogor, Bogor, Indonesia *Corresponding Author: Darniati Darniati. Laboratory of Microbiology, Faculty of Veterinary Medicine, Syiah Kuala University, Banda Aceh, Indonesia. Email: darni_zain [at] usk.ac.id Submitted: 09/09/2024 Revised: 10/06/2025 Accepted: 15/07/2025 Published: 31/08/2025 © 2025 Open Veterinary Journal
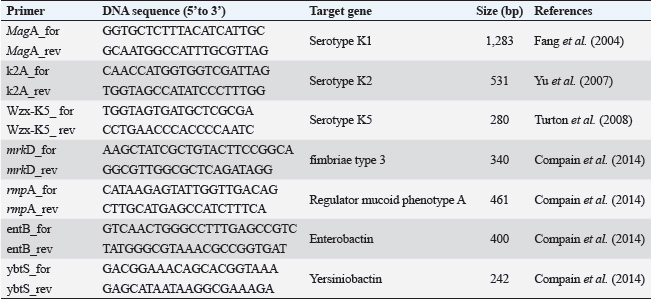
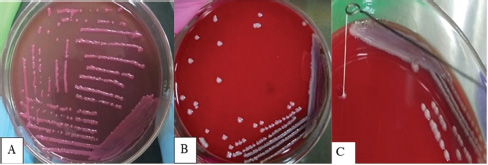
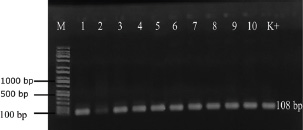
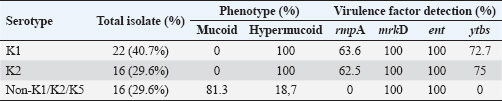
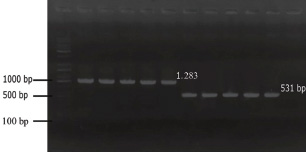
ABSTRACTBackground: Klebsiella pneumoniae is an important human and animal pathogen that causes a wide spectrum of infections. Aim: This study attempted to isolate and characterize K. pneumoniae from the respiratory tract of Aceh cattle. Methods: Pneumonic lung tissue and tracheal swabs were collected from a slaughterhouse in Banda Aceh city and the district of Aceh Besar. The isolates were identified using biochemical tests (indole, methyl red, Voges–Proskauer, sulfide indole motility, and citrate utilization) and polymerase chain reaction (PCR) to amplify the rpoB gene. The hypermucoviscous phenotype was determined by the string test. The presence of the capsule cluster genes MagA, k2A, and wzx-K5 was used to characterize the capsular serotypes K1, K2, and K5, respectively. Results: A total of 54 K. Pneumoniae isolates were obtained from 38 of 61 (62.3%) cattle. Twenty-two isolates (40.7%) were positive for the K1 genes, 16 isolates (29.6%) were positive for the K2 genes, and 16 isolates (29.6%) were negative for K1, K2, and K5 genes and classified into non-K1/K2/K5. The rmpA gene was detected in 62.5% and 63.6% of K2 and K1 isolates, respectively. mrkD and ent genes were detected in all isolates, and ytbs was detected in 72.7% and 75% of K1 and K2 isolates, respectively. Conclusion: Numerous K. pneumoniae serotypes were involved in infecting Aceh cattle, and serotype K1 had the highest prevalence. Keywords: Aceh cattle, Capsular serotype, Klebsiella pneumoniae, Pneumonic lung, Virulence genes. IntroductionKlebsiella pneumoniae is an encapsulated Gram-negative bacterium that is widely distributed in the environment. Klebsiella pneumoniae is a common opportunistic pathogen that causes infections in humans and animals (Martin and Bachman, 2018; De Koster et al., 2022). Typically, opportunistic K. pneumoniae mainly affects immunocompromised individuals or patients with pre-existing underlying diseases. Klebsiella pneumoniae is responsible for mastitis in cattle (Cheng et al., 2018); urinary tract infections (UTI) in dogs and cats (Sartori et al., 2019; Hayakawa Ito de Sousa et al., 2021); and UTI, pneumonia, meningitis, and liver abscesses in humans (Jun, 2018; Choby et al., 2020). Klebsiella pneumoniae possesses multiple virulence factors involved in the capability to initially colonize the mucosal surface. Klebsiella pneumoniae typically expresses capsular polysaccharides (CPS) and lipopolysaccharides (LPS), which are involved in mucosal colonization and the development of infections. Other K. pneumoniae virulence factors include capsules to avoid phagocytic killing, iron-scavenging systems (siderophores) for iron acquisition from the host, and fimbrial and non-fimbrial adhesions for host cell attachment (Paczosa and Mecsas, 2016; Riwu et al., 2022). There are two main pathotypes circulating worldwide: classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp). hvKp is usually associated with a hypermucoviscous K. pneumoniae strain. The hvKp exhibits large amounts of mucopolysaccharide web and extraCPS to produce thick capsules (Walker and Miller, 2020). The emergent hvKp is associated with highly invasive infections and is largely resistant to the majority of antibiotics. Klebsiella pneumoniae capsular serotypes K1, K2, K4, and K5 have been reported to cause infection in humans and animals and have a high virulence rate in experimental infections in experimental mice (Sharma et al., 2014; Jian-Li et al., 2017; M’lan_Britoh et al., 2018; Xu et al., 2022). These serotypes cause respiratory infections and septicemia more frequently than other serotypes (Jian-Li et al., 2017). On the other hand, K. pneumoniae causes pneumonia and mastitis with poor prognosis in cattle (Kot and Witeska, 2024). The invasion of hvKp in domestic animals poses a threat to livestock production and risk for public health, due to their potential role as reservoirs of multidrug-resistant K. pneumoniae strains (Cheng et al., 2018; Effah et al., 2020). Accurate pathogen detection and characterization are important for preventing controlling and managing infectious diseases in livestock, including those in Aceh cattle. However, studies related to this topic in local Indonesian cattle breeds, mainly Aceh cattle, have been poorly investigated. Aceh cattle are one of four native Indonesian cattle breeds (Aceh, Pesisir, Madura, and Bali cattle), as stipulated in the decision of the Indonesian Minister of Agriculture No: 2907/Kpts/OT/140/6/2011. The cattle have an original geographical distribution in Aceh Province and are the most widely cultivated by Aceh farmers. Therefore, this study aimed to explore the characteristics and prevalence of K. pneumoniae in Aceh cattle in Indonesia. Materials and MethodsSample collectionA total of 61 pneumonic lung tissues and tracheal swabs were collected from Aceh cattle. The samples were collected from slaughterhouses in Banda Aceh city and Aceh Besar districts, Indonesia. The tracheal swab was then cultured in a sterile Brain Heart Infusion Broth (BHIB) (Oxoid, UK) as an enrichment medium, while pneumonic lung tissue was crushed with a sterile pestle before culturing in BHIB for 24 hours at 37°C. Each sample was cultured on MacConkey agar (Oxoid, UK) and Blood agar media (Oxoid, UK) and incubated at 37°C for 24 hours. Biochemical identificationBiochemical tests were performed to determine the phenotypic characteristics of the bacterial isolates. The principal tests used for this purpose are catalase, indole, urease, methyl-red, Voges–Proskauer (VP), citrate utilization, sulfide indole motility, hydrogen sulfide production on TSI agar, and carbohydrate fermentation test (glucose, lactose, sucrose, and mannitol). PCR-based identificationSuspected colonies were identified by detecting the rpoB gene (F-CAACGGTGTGGTTACTGACG and R-TCTACGAAGTGGCCGTTTTC), which is 108 base pairs in size. We extracted nucleic acids from each colony using the boiling method according to Chen et al. (2011) and Li et al. (2022). The polymerase chain reaction (PCR) was carried out in a total volume of 25 µl consisting of template DNA (1 µl), 0.6µl of each primer (10 µM), 12.5 µl master mix MyTaq HS Mix Red (Bioline), and 10.3 µl dH2O. Amplification was performed in a Bio-Rad thermal cycler (BioRad Laboratories, Inc., Hercules, CA, USA) using the following amplification conditions: initial denaturation at 95°C for 3 minutes; 30 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 90 seconds, extension at 72°C for 90 seconds; and a final extension at 72°C for 10 minutes. The PCR products were separated by 1.5% (wt/v) agarose gel electrophoresis for 30 minutes at 120 V and visualized using a UV transilluminator. Hypermucoviscous virulent phenotype detection (string test)A string test was used to identify the hypermucoviscous phenotype of K. pneumoniae. The hvKP phenotype strains were defined by the formation of a mucoid string >5 mm in length when bacterial colonies on the blood agar plate are stretched vertically by an inoculation loop (Chang and Ong 2022; Yan et al., 2024). Capsular serotype and gene virulence amplificationA multiplex PCR assay was designed to detect MagA, k2A, and wzx-K5 genes for K1, K2, and K5 serotypes, respectively, and virulence factors (rmpA, mrkD, ent, and ytbs) gene, using specific primers (Table 1). The thermal profile for the multiplex PCR reaction was as follows: initial denaturation at 95°C for 3 minutes, followed by 30 cycles of denaturation at 95°C for 90 seconds, annealing at 55°C for 60 seconds, extension at 72°C for 60 seconds, and a final extension at 72°C for 10 minutes. The amplicons were separated at 120 V for 30 minutes in a 1.5% (wt/v) agarose gel containing ethidium bromide. Ethical approvalThe animal ethics committee of the Animal Care and Use Committee of Research and Community Service, Bogor Agricultural University, approved the present study, with ethical approval number: 144/KEH/SKE/VII/2019. ResultsPhenotypic characterization of K. pneumoniaeA total of 54 K. pneumoniae isolates were identified in this study, originating from pneumonic lung tissue and tracheal swabs of 38 Aceh cattle. Sixteen cattle were infected with different isolates in the lungs and trachea. Based on the phenotypic characteristics, K. pneumoniae was observed to be rounded, measuring 4–5 mm, and mucoid, producing pink discoloration on MacConkey agar (Fig. 1a), and whitish-gray colonies with γ-hemolysis on Blood agar (Fig. 1b). The cells were found to be Gram-negative bacilli or coccobacilli, non-spore forming, and surrounded by a thick capsule. The isolates were positive for catalase, urease production, citrate utilization, and VP. The isolates tested negative for methyl red, indole production, motility test, and H2S production. The rpoB analysis confirmed the isolates as K. pneumoniae (Fig. 2). Hypermucoviscosity and serotypes of K. pneumoniae isolatesA string test-positive K. pneumoniae was defined as a hypermucoviscous colony. In this study, 75.9% (41/54) of the K. pneumoniae isolates were string test-positive. Hyperviscosity of K. pneumoniae was observed with string formation (>5 mm) on a single colony (Fig. 1c). Serotype characterization using PCR showed that 22 (40.7%) isolates carried the MagA gene and were associated with serotype K1, 16 (29.6%) carried the k2A gene and were associated with serotype K2, and 16 (29.6%) lacked the MagA, k2A, and K5 genes and were grouped as non-K1/K2/K5 (Fig. 3). Based on the string test and molecular serotyping, isolates K1 and K2 showed hypermucoid characteristics, and only three isolates of Non-K1/K2/K5 possessed similar characteristics. Table 1. Primer set to detect the virulence factor of K. pneumoniae.
Fig. 1. Phenotypic characteristics of isolates grown on agar medium. Pink mucoid colonies on MacConkey agar (A), whitish-gray colonies on blood agar (B), and formation of viscous strings measuring >5 mm (C). Detection of virulence-associated genes in K. pneumoniae isolatesVirulence factors, such as rmpA, mrkD, ent, and ytbs, were analyzed using a multiplex PCR assay. Table 2 shows the products of the PCR assay. The results showed that the rmpA gene detected in K1 and K2 isolates was 63.6% (n=22) and 62.5% (n=16), respectively. The mrkD and ent genes were detected in all isolates, including Non-K1/K2/K5, while ytbs was detected in 72.7% (n=22) and 75% (n=16) of K1 and K2 isolates, respectively.
Fig. 2. PCR amplification of the rpoB gene for Klebsiella pneumoniae in gel electrophoresis. M: marker, Lane 1-10: K. pneumoniae isolates from pneumonic Aceh cattle. Table 2. Prevalence of capsular types and virulence factors of K. pneumoniae isolated from pneumonic Aceh cattle.
Fig. 3. Molecular typing of K. Pneumoniae capsular antigens. M: Marker. Lane 1–5: K1 Serotype (MagA), Lane 6–10: K2 serotype (k2A). DiscussionKlebsiella pneumoniae is an important opportunistic pathogen that causes a variety of infectious diseases in humans, animals, and plants, especially in immunocompromised hosts (Huang et al., 2016; Liu et al., 2023). The emergence of hypervirulent strains increases the occurrence of infection in healthy and immunocompetent hosts (Shon et al., 2013; Zhuo et al., 2025). The virulence factors of the pathogen and the decrease in mucosal immunity can increase the colonization rates of K. pneumoniae in the lungs (Teo et al., 2024). In the present study, K. pneumoniae was isolated from the infected lungs of Aceh cattle exhibiting hyperemia, congestion, consolidation, and atelectasis in the interstitial region. Klebsiella pneumoniae as the causative agent in the pneumonic lungs of Aceh cattle has been confirmed through molecular and immunohistochemistry studies in bovine pneumonic lung tissues (Darniati et al., 2021a). The presence of the rpoB gene was confirmed in all 54 K. pneumoniae isolates. The rpoB gene encodes an RNA polymerase subunit that plays an important role in transcription and contains a conserved region that is effective for identifying bacteria and as a locus for phylogenetic analysis (Chander et al., 2011). Amplification of the rpoB gene has been demonstrated to be more sensitive and discriminatory for detecting K. pneumoniae species than the 16S rRNA gene and can be used to identify organism levels at the species and subspecies (He et al., 2016). Although K. pneumoniae was a part of the normal flora in the upper respiratory system, the pathogen has been reported capable of infecting the bovine lower respiratory system (Darniati et al., 2021b). However, the virulence factors, genetic, and phenotypic variations among K. pneumoniae isolated from pneumonic lungs of cattle in Indonesia have not been widely reported (Darsana et al., 2015). In this study, K. pneumoniae was isolated from 62.3% (38/61) individuals. Positive samples were obtained from cattle showing symptoms of depression (cachexia) with or without respiratory symptoms observed before the animal was slaughtered (antemortem observations). Klebsiella pneumoniae isolates showed mucoid, non-hemolytic, and whitish-gray colonies on blood agar and formed pink mucoid colonies (lactose fermenter) on MacConkey Agar media. The results of the microviscosity test with the string test showed that 41 (75.9%) bacterial isolates had hypermucoid characters, including K1 and K2 serotypes, and 3 isolates of non-K1/K2/K5. Capsule plays a dominant role in K. pneumoniae virulence and pathogenicity. Hypercapsule production provides a more robust defense against the host’s immune system compared with non-encapsulated K. pneumoniae (Dai and Hu, 2022). In general, K. pneumoniae is classified into two pathotypes, namely hypervirulent strains (hvKp) and classic strains (cKp) based on their clinical and phenotypic characteristics (Pomakova et al., 2012). The classical K. pneumoniae exhibits high resistance to a wide range of antibiotics and causes severe pneumonia, especially in patients with chronic alcoholism, chronic obstructive pulmonary disease, and diabetes (Paczosa and Mecsas, 2016). Hypervirulent K. pneumoniae was first described in Taiwan in the mid-1980s and 1990s for causing pyogenic liver abscesses (Liu et al., 1986; Wang et al., 1998). This new variant of Klebsiella can cause serious infections in healthy individuals and infect various organs, such as the lungs, pleura, urinary tract, abdominal cavity, blood, skin, eyes, and central nervous system (Chen et al., 2010; Pomakova et al., 2012; Shon et al., 2013; Prokesch et al., 2016; Martin and Bachman, 2018). Capsular characterization of K. pneumoniae isolates from the respiratory tract of Aceh cattle showed that bacteria with hypermucoid characteristics had MagA or k2a genes. The MagA gene was detected in 22 (40.7%) isolates, and k2A was identified in 16 (29.6%%) isolates. The wzx-K5 gene specific to the K5 serotype was not detected in the respiratory tract of Aceh cattle. Klebsiella pneumoniae serotype K1 was detected by PCR with the target gene MagA located in the operon specific for serotype K1 and yielded a fragment measuring 1.283 bp, matching the size obtained by Turton et al. (2008). The K2 serotype was detected by targeting the k2A gene and yielded a fragment size of 543 bp, matching the size obtained by Yu et al. (2008). Non-K1/K2/K5 isolates generally had lower viscosity than K1 and K2 isolates, although 3 isolates showed a string length of ± 5 mm in this study. Although capsular serotypes K1 and K2 are often associated with hvKp, cKp can also have a hypermucoid character (Shon et al., 2013). Evolutionary mechanisms contribute to the emergence of identical K-types, even in the absence of phylogenetic relationships between the serotypes (Brisse et al., 2009). In this study, the K1 serotype was more predominant than the K2 and non-K1/K2/K5. Although serotype K1 is considered the most virulent strain of K. pneumoniae, capsular serotypes K1 and K2 are associated with hvKp, and major capsular serotypes are involved in liver abscess and metastatic septic complications (De Francesco et al., 2023). In the current study, the prevalence of capsular serotypes K1 and K2 was approximately 70.4%. This is higher than the study by Mario et al. (2023), which identified a prevalence of hvKp at 24.2% in Giza Governorate, Egypt (Mario et al., 2023). Previous studies have reported that K1 and K2 strains have higher virulence than other serotypes (Wang et al., 2021). The presence of thick polysaccharide capsules at the cell surface of K1 and K2 serotypes directly or indirectly contributes to anti-phagocytic activity (Lee et al., 2014; Opal, 2014; Al Ismail et al., 2025). Capsules could protect K. pneumoniae by inhibiting host immune cell phagocytosis, preventing early immune response activation, and hampering lysis by complement and antimicrobial peptides. Capsule confers resistance to opsonophagocytosis in K. pneumoniae and contributes to biofilm formation (Dai and Hu, 2022). Macrophage lectin receptors initiate phagocytosis through the recognition of mannose or rhamnose sugar patterns on the microbial capsule (Xu et al., 2024). On the other hand, K1 and K2 capsule types lack these repeated sugars, hence facilitating the evasion from lectinophagocytosis and macrophage killing (Athamna et al., 1991; Kabha et al., 1995, Pan et al., 2015). Moreover, K1 isolates generally express the fba gene in high glucose concentrations, causing inhibition of neutrophil-mediated phagocytosis and intracellular killing (Lee et al., 2020). Sialic acid (Sia) is a component of CPS in the hvKp phenotype that is directly or indirectly responsible for anti-phagocytic activity (Lee et al., 2014; Opal, 2014). Virulence factors of K. pneumoniae play a critical role in determining its prevalence and pathogenicity within the host. Several virulence factors, including capsules, LPS, siderophores, and fimbriae, have been identified in K. pneumoniae. These factors play an important role in the existence of pathogens in the environment and host (Paczosa and Mecsas, 2016). In this study, the virulence factor of capsular serotype K1, K2, and non-K1/K2/K5 characterization was performed to detect the presence of regulator of mucoid phenotype A (rmpA), type 3 fimbrial adhesin gene (mrkD), enterobactin (ent), and yersiniabactin (ytbs). Molecularly, there are two genes in K. pneumoniae that are associated with invasive infections, namely mucoviscosity-associated gene A (MagA) and mucoid regulator phenotype A (rmpA) (Lin et al., 2020). The rmpA gene is a plasmid-carried gene and plays a role in capsular polysaccharide synthesis and enhances the hypermucoviscus phenotype (Compain et al., 2014). In this study, the rmpA gene was detected in 62.5% and 63.6% of the K2 and K1 serotypes, respectively. The pathogenicity of K. pneumoniae is determined by the ability of the pathogen to secrete multiple types of siderophores to scavenge ferric iron from the environment. Hypervirulent K. pneumoniae strains express four siderophores: enterobactin, yersiniabactin, salmochelin, and aerobactin (Miethke and Marahiel, 2007; Bachman et al., 2012). Enterobactin (ent) has the highest affinity for iron of any molecule and is carried by almost all cKp and hvKp strains (Lawlor et al., 2007). In this study, enterobactin was detected in all isolates of K. pneumoniae. In addition, yersinibactin was detected in approximately 72.7% and 75% of K1 and K2 serotypes, respectively. Yersiniabactin is an important virulence factor in cKp and promotes respiratory tract infections (Lawlor et al., 2007; Miethke and Marahiel, 2007). Yersiniabactin was detected in approximately 18% of the classical isolates and 90% of the hypervirulent isolates (Bachman et al., 2011). Lipocalin-2 cannot neutralize yersiniabactin in the lungs, thus increasing the risk of infection in the lungs. However, yersiniabactin is unable to access the iron required for K. pneumoniae growth in the presence of the host protein transferrin in the blood. Thus, isolates that only produce yersiniabactin will not be capable of disseminating to the blood (Bachman et al., 2011). Fimbrial adhesins enable K. pneumoniae to adhere to host cells, facilitating a critical step in the initiation of host invasion. Type 1 and type 3 fimbriae mediate adhesion factors in K. pneumoniae. Type 1 and 3 fimbriae are detected in almost all K. pneumoniae strains to mediate cell attachment and promote biofilm formation. Biofilms are useful for evading host defense mechanisms, antimicrobials, and disinfectants (Johnson, 2011). The mrkABCD gene cluster encodes type 3 fimbriae (Tarkkanen et al. 1998). The mrkA subunit is the largest structure of this gene, with the adhesion factor mrkD at the end of mrkA. Type 3 fimbriae mediate Klebsiella adhesions to tracheal cells, tongue cells, and lungs via mrkD (Hornick et al., 1992; Sonbol et al., 2013). MrkD gene can be detected in all K1, K2, and non-K1/K2/K5 isolates. Although K1 and K2 are considered the most virulent serotypes, clinical and epidemiological studies have shown that some non-K1/K2/K5 serotypes can also cause various infections in humans and exhibit high mortality rates (Qu et al., 2015; Yao et al., 2015; Yu et al., 2015). Therefore, the presence of these serotypes can still pose a risk to animal and human health. According to Paczosa and Mecsas (2016), cKp infection generally occurs in hosts who are immunosuppressed or have immune response disorders. Decreased immune response in the host can be caused by heat stress, injury, fatigue, dehydration, and nutritional deficiencies in the process of weaning and livestock transportation (McGill and Sacco, 2020). Extensive smallholder farming conditions, hot environment/climate, and limited nutrition have the potential to cause animals to experience prolonged stress or depression, which triggers a decrease in immune response, making them more easily infected with various diseases, including K. pneumoniae. This statement supports the results of studies showing a fairly high prevalence of K. pneumoniae in the respiratory tract of extensively reared Aceh cattle. In conclusion, K. pneumoniae is among the pathogens potentially causing lower respiratory infection in Aceh cattle. The proportion of Hypermucoviscous strain isolates among K. pneumoniae-positive cultures is higher than that of low-mucoid strains, indicating a high prevalence of hvKP. Identified hvKp underscores the need for closer surveillance of livestock populations. Effective surveillance requires comprehensive data on the genetic diversity, antimicrobial resistance mechanisms, and transmission dynamics of these isolates. Molecular studies comparing isolates from animals and humans in the same region are essential for assessing zoonotic potential. In addition to controlling the spread of isolated K. pneumoniae, it is strongly recommended to improve biosecurity at the farm level, implement targeted vaccination strategies, and enforce responsible antibiotic use policies. AcknowledgmentsThe authors would like to thank IPB University and Universitas Syiah Kuala, Indonesia, for their support during this research. Conflict of interestThe authors declare no conflict of interest. Authors’ contributionsDN, SS; Methodology, validation, and analysis. DN, EH; Data curation and writing-original draft preparation. DN, SS, EH; Executed the analysis, interpreted the data, and revised the manuscript. Data availabilityAll data supporting the findings of this study are available within the manuscript. ReferencesAl Ismail, D., Campos-Madueno, E.I., Donà, V. and Endimiani A. 2025. Hypervirulent Klebsiella pneumoniae (hvKp): overview, epidemiology, and laboratory detection. Pathog. Immun. 10(1), 80–119. Athamna, A.B.E.D., Ofek, I.T.Z.H.A.K., Keisari, Y., Markowitz, S., Dutton, G.G. and Sharon, N. 1991. Lectinophagocytosis of encapsulated Klebsiella pneumoniae mediated by surface lectins of guinea pig alveolar macrophages and human monocyte-derived macrophages. Infect. Immun. 59, 1673–1682. Bachman, M., Lenio, S., Schmidt, L., Oyler, J.E. and Weiser J.N. 2012. Interaction of lipocalin 2, transferrin, and siderophore determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 3, 1–11. Bachman, M.A., Oyler, J.E., Burns, S.H., Caza, M.l., Le´pine, F.O., Dozois, C.M. and Weiser, J.N. 2011. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 79, 3309–3316. Brisse, S., Fevre, C., Passet, V., Issenhuth-Jeanjean, S., Tournebize, R., Diancourt, L. and Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4, e4982. Chander, Y., Ramakrishnan, M.A., Jindal, N., Hanson, K. and Goyal S.M. 2011. Differentiation of Klebsiella pneumoniae and K. oxytoca by multiplex polymerase chain reaction. Int J Appl Res inVeterinary Med. 9, 138–142. Chang, C.Y. and Ong, E.L. 2022. Positive string test in hypervirulent Klebsiella pneumoniae liver abscess. Oxf. Med. Case Rep. 4, 143–144. Chen, K.J., Hwang, Y.S., Wang, N.K. and Chao, A.N. 2010. Endogenous Klebsiella pneumoniae endophthalmitis with renal abscess: report of two cases. Int J Infect Dis. 14(5), e429–e432. Chen, L., Mediavilla, J.R., Endimiani, A., Rosenthal, M.E., Zhao, Y., Bonomo, R.A. and Kreiswirth, B.N. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J. Clin. Microbiol. 49(2), 579–585. Cheng, F., Li, Z., Lan, S., Liu, W., Li, X., Zhou, Z., Song, Z., Wu, J., Zhang, M. and Shan, W. 2018. Characterization of Klebsiella pneumoniae associated with cattle infections in southwest China using multilocus sequence typing (MLST), antibiotic resistance, and virulence-associated gene profile analysis. Braz. J Microbiol. 49(2018), 93–100. Choby, J.E., Howard-Anderson, J. and Weiss, D.S. 2020. Hypervirulent Klebsiella pneumoniae-clinical and molecular perspectives. J. Intern. Med. 287, 283–300. Compain, F., Babosan, A., Brisse, S., Genel, N., Audo, J., Ailloud, F., Kassis-Chikhani, N., Arlet, G. and Decréa, D. 2014. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. ASM J. 52, 4377–4380. Dai, P. and Hu, D. 2022. The making of hypervirulent Klebsiella pneumoniae. J. Clin. Lab. Anal. 36(12), 1–16. Darniati, D., Setiyaningsih, S., Agungpriyono, D.R. and Handharyani, E. 2021a. Development of monospecific polyclonal antibodies against hypervirulent Klebsiella pneumoniae. Biodiversitas 22, 99–105. Darniati, D., Setiyaningsih, S., Agungpriyonom, D.R. and Handharyani, E. 2021b. First evidence of Klebsiella pneumoniae infection in Aceh cattle: pathomorphology and antigenic distribution in the lungs. Vet World 14, 1007–1013. Darsana, I.G.O., Dibia, I.N. and Mahatmi, H. 2015. Detection of Mycobacterium bovis and Klebsiella pneumoniae at bali cattle slaughterhouse by culture analysis and PCR. J. Ilmu Kesehatan Hewan 3(2), 51–54. De Francesco, M.A., Tiecco, G., Scaltriti, E., Piccinelli, G., Corbellini, S., Gurrieri, F., Crosato, V., Moioli, G., Marchese, V., Focà, E. and Bertelli, D.A. 2023. First Italian report of a liver abscess and metastatic endogenous endophthalmitis caused by ST-23 hypervirulent Klebsiella pneumoniae in an immunocompetent individual. Infection 51(1), 271–276. De Koster, S., Rodriguez Ruiz, J.P., Rajakani, S.G., Lammens, C., Glupczynski, Y., Goossens, H. and Xavier, B.B. 2022. Diversity in the characteristics of Klebsiella pneumoniae ST101 of human, environmental, and animal origin. Front. Microbiol. 13, 838207. Effah, C.Y., Sun, T., Liu, S. and Wu, Y. 2020. Klebsiella pneumoniae: an increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 19, 1–9. Fang, C.T., Chuang, Y.P., Shun, C.T., Chang, S.C. and Wang, J.T. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199, 697–705. Hayakawa Ito de Sousa, A.T., dos Santos Costa, M.T., Makino, H., Cândido, S.L., de Godoy Menezes, I. and Lincopan, N. 2021. Multidrug-resistant Mcr-1 gene-positive Klebsiella pneumonia St307 causing urinary tract infection in a cat. Brazil J Microbiol. 52, 1043–1046. He, Y., Guo, X., Xiang, S., Li, J., Li, X., Xiang, H., He, J., Chen, D. and Chen, J. 2016. Comparative analyses of phenotypic methods and 16S rRNA, khe, rpoB genes sequencing for of clinical isolates of Klebsiella pneumoniae. Antonie Van Leeuwenhoek 109, 1029–1040. Hornick, D., Allen, B., Horn, M. and Clegg, S. 1992. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect Immun. 60, 1577–1588. Huang, M., Li, L.I.N., Wu, Y.X., Honhing, H., He, P.F., Li, G.Z., He, P.B., Xiong, G.R., Yuan, Y.U.A.N. and He, Y.Q., 2016. Pathogenicity of Klebsiella pneumonia (KpC4) infecting maize and mice. J. Integr. Agric. 15, 1510–1520. Jian-Li, W., Yuan-Yuan, S., Shou-Yu, G., Fei-Fei, D., Jia-Yu, Y., Xue-Hua, W., Yong-Feng, Z., Shi-Jin, J. and Zhi-Jing, X. 2017. Serotype and virulence genes of Klebsiella pneumoniae isolated from mink and its pathogenesis in mice and mink. Sci Rep. 7, 1–7. Johnson, J.G. 2011. Regulation of type 3 fimbrial gene expression in Klebsiella pneumoniae. PhD Dissertation. University of IOWA, IOWA. Jun, J.B. 2018. Klebsiella pneumoniae liver abscess. Infect. Chemother. 50, 210–218. Kabha, K., Nissimov, L., Athamna, A., Keisari, Y., Parolis, H., Parolis, L., Grue, R., Schlepper-Schafer, J., Ezekowitz, A., Ohman, D. and Ofek I. 1995. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun. 63, 847–852. Kot, B. and Witeska, M. 2024. Review of antimicrobial resistance of Klebsiella pneumoniae isolated from poultry, cattle and pigs. Animal 18, 101345. Lawlor, M.S., O’connor, C. and Miller, V.L. 2007. Yersiniabactin is a virulence factor Klebsiella pneumoniae during pulmonary infection. Infection and immunity. J Immunol. 75, 1463–1472. Lee, C.H., Changm, C.C., Lium, J.W., Chen, R.F. and Yang, K.D. 2014. Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence 5, 673–679. Lee, C.H., Chuah, S.K., Chang, C.C. and Chen, F.J., 2020. The surface protein fructose-1, 6 bisphosphate aldolases of Klebsiella pneumoniae serotype K1: role of interaction with Neutrophils. Pathogens 9, 1009. Li, N., Wangm L., Wang, F., Chen, H., Tao, S., Zhu, Q., Liu, L., Liang, W. and Ma, F. 2022. Rapid detection of Klebsiella pneumoniae carrying virulence gene rmpA2 by recombinase polymerase amplification combined with lateral flow strips. Front. Cell. Infect. Microbiol. 12(2), 1–10. Lin, Z.W., Zheng, J.X., Bai, B., Xu, G.J., Lin, F.J., Chen, Z., Sun, X., Qu, D., Yu, Z.J. and Deng, Q.W. 2020. Characteristics of hypervirulent Klebsiella pneumoniae: does low expression of rmpA contribute to the absence of hypervirulence? Front. Microbiol. 11, 436. Liu, Y., Huang, L., Cai, J., Zhu, H., Li, J., Yu, Y., Xu, Y., Shi, G. and Feng, Y. 2023. Clinical characteristics of respiratory tract infection caused by Klebsiella pneumoniae in immunocompromised patients: a retrospective cohort study. Front. Cell. Infect. Microbiol. 13, 1137664. Liu, Y.C., Cheng, D.L. and Lin, C.L. 1986. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med 146, 1913–1916. M’lan-Britoh, A., Meité, S., Boni, C., Zaba, F., Koffi, K., Guessennd, N., Kakou, N., Kette, H.F. and Dosso, M. 2018. First molecular investigation of capsular serotyping and hypervirulent (hvkp) of K. pneumoniae in University Hospital Center of Yopougon Cote D’ivoire. Afr. J. Cln. Exper. Microbiol. 19, 70–75. Mario, E., Hamza, D. and Abdel-Moein, K. 2023. Hypervirulent Klebsiella pneumoniae among diarrheic farm animals: a serious public health concern. Comp. Immunol. Microbiol. Infect. Dis. 102(102077), 1–7. Martin, R.M. and Bachman, M.A. 2018. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8(4), 1–15. McGill, J.L. and Sacco, R.E. 2020. The immunology of bovine respiratory disease: recent advancements. Vet. Clin. Food Anim. 36, 333–348 Miethke, M. and Marahiel, M. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. Opal, S.M. 2014. Significance of sialic acid in Klebsiella pneumoniae K1 capsules. Virulence 5, 648–649. Paczosa, M.K. and Mecsas, J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. Pan, Y.J., Lin, T.L., Chen, C.T., Chen, Y.Y., Hsieh, P.F., Hsu, C.R., Wu, M.C. and Wang, J.T. 2015. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 5, 15573. Pomakova, D.K., Hsiao, C.B., Beanan, J.M., Olson, R., MacDonald, U., Keynan, Y. and Russo, T.A. 2012. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: An emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 3, 981–989. Prokesch, B.C., TeKippe, M., Kim, J., Raj, P., Tekippe, E.M.E. and Greenberg, D.E. 2016. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 16(9), e190–e195. Qu, T., Zhou, J., Jiang, Y., Shi, K., Li, B., Shen, P., Wei, Z. and Yu, Y. 2015. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 15(1), 161. Riwu, K.H.P., Effendi, M.H., Rantam, F.A., Khairullah, A.R. and Widodo, A. 2022. A review: virulence factors of Klebsiella pneumonia as emerging infection on the food chain. Vet. World 15, 2172. Sartori, L., Sellera, F.P., Moura, Q., Cardoso, B. and Cerdeira, L. 2019 Lincopan Multidrug-resistant Ctx-M-15-positive Klebsiella pneumonia st307 causing urinary tract infection in a dog in Brazil. J. Glob. Antimicrob. Resist. 19, 96–97. Sharma, S., Sharma, P. and Shringi, B. 2014. Phenotypic and genotypic characterization of Klebsiella pneumoniae obtained from Egyptian vultures and steppes eagles from India. Isr. J. Vet. Med. 69, 123–129. Shon, A.S., Bajwa, R.P. and Russo, T.A. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. Sonbol, F.I., El-Banna, T.E., Abd El-Aziz, A.A. and Al-Madboly, L.A. 2013. Conjugative plasmid mediating adhesive pili in virulent Klebsiella pneumoniae isolates. Arch. Clin. Microbiol. 4, 1–9. Tarkkanen, A., Westerlund-Wikstrom, B., Erkkila, L. and Korhonen, T. 1998. Immunohistological localization of the MrkD adhesin in the type 3 fimbriae of Klebsiella pneumoniae. Infect. Immun. 66, 2356–2361. Teo, T.H., Ayuni, N.N., Yin, M., Liew, J.H., Chen, J.Q., Kurepina, N., Rajarethinam, R., Kreiswirth, B.N., Chen, L. and Bifani, P. 2024. Differential mucosal tropism and dissemination of classical and hypervirulent Klebsiella pneumoniae, infection. Iscience. 27, 108875. Walker, K.A. and Miller, V.L. 2020. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr. Opin. Microbiol. 54, 95–102. Wang, J.H., Liu, Y.C., Lee, S.S., Yen, M.Y., Chen, Y.S., Wang, J.H., Wann, S.R. and Lin, H.H. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26, 1434–1438. Wang, T.C., Lin, J.C., Chang, J.C., Hiaso, Y.W., Wang, C.H., Chiu, S.K., Fung, C.P., Chang, F.Y. and Siu, L.K., 2021. Virulence among different types of hypervirulent Klebsiella pneumoniae with multi-locus sequence type (MLST)-11, Serotype K1 or K2 strains. Gut Pathogens 13(1), 40. Xu, L., Li, J., Wu, W., Wu, X. and Ren, J. 2024. Klebsiella pneumoniae capsular polysaccharide: Mechanism in regulation of synthesis, virulence, and pathogenicity. Virulence 15(1), 2439509. Xu, Y., Ni, L., Guan, H., Chen, D., Qin, S. and Chen L. 2022. First report of potentially pathogenic Klebsiella pneumoniae from serotype K2 in Mollusk Tegillarca granosa and genetic diversity of Klebsiella pneumoniae in 14 species of edible aquatic animals. Foods 11(24), 4058. Yan, W., Xu, D., Shen, Y., Dong, F. and Ji, L. 2024. Molecular epidemiology of string test-positive Klebsiella pneumoniae isolates in Huzhou, China, 2020–2023. Front. Cell. Infect. Microbiol. 14(1411658), 1–10. Yao, B., Xiao, X., Wang, F., Zhou, L., Zhang, X. and Zhang, J. 2015. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 37, 107–112. Yu, W.L., Ko, W.C., Cheng, K.C., Lee, C.C., Lai, C.C. and Chuang, Y.C. 2008. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 62(1), 1–6. Yu, W.L., Lee, M.F., Tang, H.J., Chang, M.C. and Chuang, Y.C. 2015. Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum β-lactamase-producing Klebsiella pneumoniae isolates. Virulence 6(2), 162–172. Yu, W., Fung, C., Ko, W., Cheng, K., Lee, C. and Chuang, Y. 2007. Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis 195(8), 1235–1236. Zhuo, X., Lei, Z., Pu, D., Wu, Y., Zhao, J. and Cao, B. 2025. Hypervirulent Klebsiella pneumoniae have better clinical outcomes than classical Klebsiella pneumoniae for lower respiratory tract infection patients. BMC Microbiol. 25(1), 40. | ||
| How to Cite this Article |
| Pubmed Style Darniati D, Handharyani E, Setiyaningsih S. Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle. Open Vet. J.. 2025; 15(8): 3737-3745. doi:10.5455/OVJ.2025.v15.i8.38 Web Style Darniati D, Handharyani E, Setiyaningsih S. Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle. https://www.openveterinaryjournal.com/?mno=219503 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i8.38 AMA (American Medical Association) Style Darniati D, Handharyani E, Setiyaningsih S. Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle. Open Vet. J.. 2025; 15(8): 3737-3745. doi:10.5455/OVJ.2025.v15.i8.38 Vancouver/ICMJE Style Darniati D, Handharyani E, Setiyaningsih S. Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle. Open Vet. J.. (2025), [cited January 25, 2026]; 15(8): 3737-3745. doi:10.5455/OVJ.2025.v15.i8.38 Harvard Style Darniati, D., Handharyani, . E. & Setiyaningsih, . S. (2025) Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle. Open Vet. J., 15 (8), 3737-3745. doi:10.5455/OVJ.2025.v15.i8.38 Turabian Style Darniati, Darniati, Ekowati Handharyani, and Surachmi Setiyaningsih. 2025. Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle. Open Veterinary Journal, 15 (8), 3737-3745. doi:10.5455/OVJ.2025.v15.i8.38 Chicago Style Darniati, Darniati, Ekowati Handharyani, and Surachmi Setiyaningsih. "Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle." Open Veterinary Journal 15 (2025), 3737-3745. doi:10.5455/OVJ.2025.v15.i8.38 MLA (The Modern Language Association) Style Darniati, Darniati, Ekowati Handharyani, and Surachmi Setiyaningsih. "Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle." Open Veterinary Journal 15.8 (2025), 3737-3745. Print. doi:10.5455/OVJ.2025.v15.i8.38 APA (American Psychological Association) Style Darniati, D., Handharyani, . E. & Setiyaningsih, . S. (2025) Molecular analysis of Klebsiella pneumoniae serotypes and virulence genes isolated from pneumonic Aceh cattle. Open Veterinary Journal, 15 (8), 3737-3745. doi:10.5455/OVJ.2025.v15.i8.38 |