| Research Article | ||
Open Vet. J.. 2025; 15(5): 1941-1946 Open Veterinary Journal, (2025), Vol. 15(5): 1941-1946 Research Article Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, LibyaSumayah J. Aishbani1, Huda H. Al-Griw1, Jihan A. Al-sharif 2, Elfurgani S. Karim3, Samira A. Farag2, Mohamed O. Ahmed1 and Yousef M. Abouzeed1*1Department of Microbiology and Parasitology, Faculty of Veterinary Medicine, University of Tripoli, Tripoli, Libya 2Department Food Health Control, Faculty of Veterinary Medicine, University of Tripoli, Tripoli, Libya 3Microbiology Lab Department, National Center for Animal Health, Tripoli, Libya *Corresponding Author: Yousef. M. Abouzeed. Department of Microbiology and Parasitology, Faculty of Veterinary Medicine, University of Tripoli, Tripoli, Libya. Email: y.abouzeed [at] uot.edu.ly Submitted: 04/08/2024 Revised: 15/03/2025 Accepted: 13/04/2025 Published: 31/05/2025 © 2025 Open Veterinary Journal
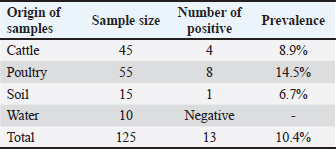
ABSTRACTBackground: Nontyphoidal Salmonella (NTS) serovars remain a potential hazard to human and animal health. Food animals with Salmonella infections can either be cured or remain asymptomatic carriers, serving as a potential source of infection to humans and other healthy animals. Salmonella may also be a reservoir of resistance determinants that can be transferred to other commensal organisms, particularly in the gastrointestinal tract. Aim: This article addresses the prevalence of Salmonella serovars in important food animals and their environment. Antibiotics susceptibility profiling of Salmonella serovars against important antibiotics used in human and animal medicines was also investigated. Methods: In total, 125 fecal samples were collected from food animals (n=100 fecal samples) and from the environment (n=25). Salmonellae were isolated using standard laboratory procedures, and typical isolates were further confirmed using laboratory biomedical testing and the automated microbial identification system VITEK 2. Confirmed Salmonella isolates were further analyzed for their susceptibility to antimicrobial agents. Results: A total of 10.4% (13/125) of samples were found positive for Salmonella by conventional isolation, biochemical identification methods, and VITEK 2 system. The prevalence of Salmonella spp. in cattle feces was 8.9% (4/45), whereas the prevalence in poultry samples was 14.5% (8/55). Only one sample from soil was found positive for Salmonella representing 6.7% (1/25). Serologically, only five Salmonella isolates were further confirmed and typed into three serovars: S. Montevideo (n=2), S. Kentucky (n=1), and S. Typhimurium (n=2). All Salmonella isolates displayed minimum inhibitory concentrations values above the susceptibility breakpoint for ampicillin, piperacillin, and second-generation cephalosporins but showed reduced susceptibility to third-generation cephalosporins. All the isolates also displayed different levels of resistance to ciprofloxacin and levofloxacin. Resistance to the aminoglycosides (amikacin, tobramycin, and gentamicin) was also rerecorded. Conclusion: The current study demonstrated that the high resistance rates to fluoroquinolones can compromise important therapeutic options against salmonellosis posing also public health concerns. Keywords: Salmonella serovars, Antibiotic susceptibility, Prevalence, Libya. IntroductionSalmonella species pose a substantial global public health concern due to their ability to cause severe disease in both humans and animals. Although salmonellae are ubiquitous in nature, the primary source of infection is the intestinal tract of infected or colonized animals and humans. In humans, infection is often associated with the consumption of contaminated foods of animal origin (EFSA and ECDC, 2017). Salmonella is a Gram-negative bacterium that belongs to the Enterobacteriaceae family, consisting of over 2,500 different serotypes. Additionally, Salmonella is a resilient bacterium that can endure dry conditions for weeks and remain submerged in water for months. Salmonella serotypes can cause disease in humans, with some species being host-specific and others present in a wide range of hosts (WHO, 2020). While most of the species cause uncomplicated gastroenteritis, some can be life-threatening, especially in vulnerable and immunocompromised individuals (Kurtz et al., 2017). Salmonella enterica serotypes Enteritidis and Typhimurium are the primary serotypes transmitted from animals to humans; however, other species are increasingly reported with variable epidemiological distribution worldwide (WHO, 2020). While antibiotic treatment is generally unnecessary for uncomplicated Salmonella gastroenteritis, recent studies indicate that a brief course of antibiotics, such as ceftriaxone, may aid clinical recovery in severe cases. Nevertheless, antimicrobial therapy is necessary in case of invasive infections, such as bacteraemia, osteomyelitis, and meningitis (Chen et al., 2013). However, the use and misuse of antimicrobial drugs in human and animals, including for prophylactic and growth-promoting purposes in food animals, are linked to the widespread dissemination of antimicrobial resistance in Salmonella and other pathogens mainly attributed to mobile genetic elements (Bouchrif et al., 2009). Genetic and genomic changes in Salmonella have increased its virulence and drug resistance, raising public health concerns. As a result, surveillance and epidemiological studies in humans and animals are essential. Such a course may significantly aid our understanding of the mechanisms of antimicrobial resistance in salmonellae, which can aid the development of effective strategies to compact its spread. The global emergence of multidrug resistance (MDR) Salmonella species and the variability of resistance patterns from one geographic area to another have necessitated the need for information on Salmonella. Salmonellae are important zoonotic pathogens with public health significance, requiring the determination of antibiotics resistance patterns, particularly from food animals and environmental sources. Recently, a report from Libya revealed the important role of pet animals as a reservoir of multidrug-resistant Salmonella (Elnageh et al., 2021). Therefore, the aim of this study was to investigate the occurrence of Salmonella isolates from selected food animals and environmental sources and to determine the susceptibility of these isolates to commonly used antibiotics in humans and veterinary medicine. Materials and MethodsCriteria of samplingA total of 125 samples were collected in this study from five farms located in the eastern area of Tripoli during the period 2017–2018. Samples included 100 fecal samples from food animals (45 from cattle and 55 from poultry) and 25 environmental samples (15 from soil and 10 from water sources) (Table 1). Samples from animals were collected on the basis that animals were healthy and showed no signs of clinical manifestations. Samples from the farm’s environment were collected by random selection from soil and water sources. Collection of samplesFecal samples were collected from animals following strict sterile conditions following proper handling procedure. Approximately, 5 g of fecal rectal sample from each animal was collected and a swab fecal samples from 5-week-old broiler cobb chickens were collected in a sterile suitable container and kept in cool place. Soil and water samples were also obtained using sterile container from each included farm. Samples were sent to the Department of Microbiology and Parasitology, Faculty of Veterinary Medicine, University of Tripoli, for initial processing within 4 hours. Isolation and laboratory identification of salmonellae were performed at the laboratory of Animal Health Centre in Tripoli, Libya. Table 1. Prevalence of Salmonella isolates from food animals and environmental sources.
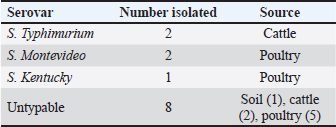
Processing of samplesApproximately 3 g of fecal sample from cattle and the cotton tip of swab sample from poultry were each mixed in 5 ml of sterile water in a bijoux tube and mixed using a vortex to obtain the homogenized sample. One milliliter of fecal suspension was added to 10 ml of buffered peptone water and incubated aerobically at 37°C for 24 hours. Afterward, 1 ml was transferred to 10 ml of Rappaport-Vassiliadis broth and incubated aerobically at 42°C for 24 hours. Similarly, 3 ml of water and 2 g of soil samples were added to 10 ml of buffered peptone water and incubated at 37°C for 24 hours. All incubated broth and mediums were inoculated onto nutrient agar and incubated at 37°C for 24 hours. Based on the cultural characteristics of the overnight-grown cultures, only one typical isolated colony representing a single sample was selected and further subjected to identification and characterization tests. The single typical colonies were then transferred and overnight grown on nutrient agar and then subjected to Gram stain and lactose fermentation test. Definite and confirmed Salmonella isolates were sent to the Animal Health Centre to fully characterize to Salmonella serovars. Salmonella strains were serotyped according to the Kauffmann–White scheme using the slide agglutination test to identify somatic O and flagellar H antigens. An in-house Salmonella strain served as a positive control during the isolation and identification process. Definite characterization and antimicrobial susceptibility testingThe Vitek 2 system (bioMérieux) is a fully automated identification and antimicrobial susceptibility testing system that accommodates 60 identification or susceptibility cards at one time in one module. The instrument is widely used for bacterial identification and susceptibility testing. The system is designed to decrease the turnaround time for the identification of bacteria and determination of antimicrobial susceptibilities. The adopted procedure was performed following the protocol published by O’Hara and Miller, 2003. ResultsPrevalence of Salmonella strains from food animals and environmental samplesSalmonella was detected in 10.4% (n=13/125) of the tested samples. Among the samples of each source, 8.9% (n=4/45) were from cattle fecal samples and 14.5% (n=8/55) were from poultry samples, whereas only one environmental sample from soil was found positive, 6.7% (n=1/25) for Salmonella. All water samples were negative for Salmonella (Table 2). Table 2. Salmonella serovars identified from food animals and environmental sources.
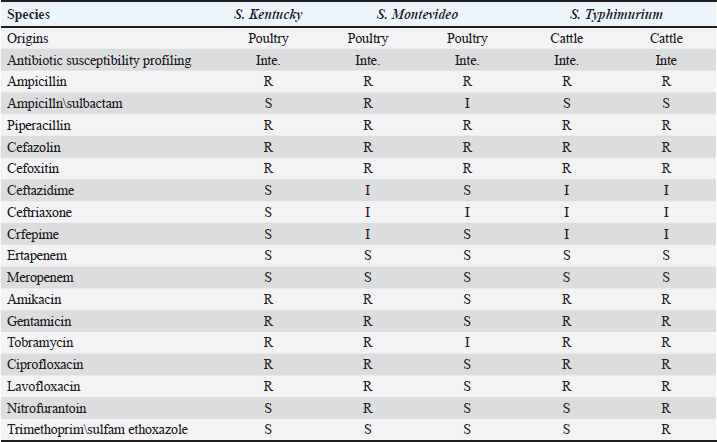
Table 3. Antibiogram of Salmonella isolates according to their original sources (typable isolates n=5).
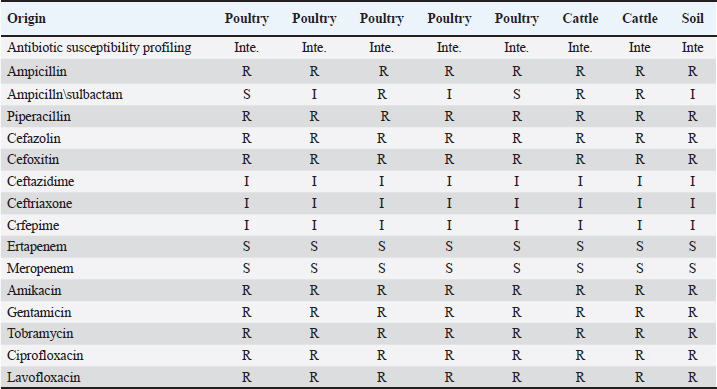
Table 4. Antibiogram of Salmonella isolates according to their original sources (untypable isolates n=8).
The identified Salmonella isolates were further confirmed serologically and characterized using the VITEK 2 system into three serovars, S. Montevideo (n=2), S. Kentucky (n=1), and S. Typhimurium (n=2), and eight isolates were found to be untypable. The susceptibility of Salmonella isolates was evaluated using the VITEK 2 system based on minimum inhibitory concentration (MIC) values as detailed in Tables 3 and 4. DiscussionIn this study, Salmonella was identified in 10.4% of samples with a prevalence of 8.9% in the tested cattle samples, of which two samples were identified as positive for S. typhimurium. S. typhimurium strains were frequently isolated from apparently healthy cattle, playing an important role as the main reservoir for human infection (Ghoddusi et al., 2019). This serovar has been implicated in many human Salmonella outbreaks associated with consumption of undercooked or ground beef. Salmonella typhimurium is a globally prevalent serovar largely associated with MDR phenotype (Abouzeed et al., 2000; Glenn et al., 2011). S. typhimurium isolated in this study expressed an MDR phenotype, including important antibiotic classes. Previous study conducted at the Health Centre in Brack town in Southwestern Libya revealed that the prevalence of human Salmonella was 17.5% in tested samples and infection was mostly reported in young children, displaying low level of resistance to many antibiotics (Altayyar et al., 2016). Poultry are often asymptomatic carriers of various Salmonella serovars that constitute the main source of human salmonellosis worldwide (Mshelbwala et al., 2017). In this study, the prevalence of Salmonella isolates in poultry fecal swabs was 14.5%, which was relatively higher than that of the cattle. Serotyping of the poultry isolates revealed that two isolates were S. Montevideo: one isolate was identified as S. Kentucky and the others were untypable. Indeed, S. Montevideo is commonly associated with human Salmonella outbreaks in which the infection is usually traced back to poultry and their products. This serovar is known to be potentially pathogenic to human. Salmonella Montevideo was reported to be the most frequently reported serovar in North America (Gutema et al., 2019). Many outbreaks reported in US during 2012 were linked to live poultry (CDC, 2010). S. Montevideo can be easily isolated from cecal and cloacal swabs of infected chickens, aligning with the current study’s findings. In the current study, the serovar S. Kentucky was encountered among the poultry isolates and found to be expressing the MDR phenotype. This serovar has been implicated in food poisoning in the EU (EFSA and ECDC, 2017). A study by Bouzidi and colleagues has associated human infection with S. Kentucky to poultry and poultry meat (Bouzidi et al., 2011). Another study reveals a case of S. enterica serovar Kentucky of sequence type 198 (ST198) that produced OXA-48 and isolated from perianal screening cultures of a patient who was transferred from Libya to Switzerland. This highlights the need for early and accurate screening methods to stop the spread of carbapenemase producers imported from endemic areas (Seiffert et al., 2014). Antibiotics resistance in pathogenic bacteria of animal and human origin is a great public health concern. Several epidemiological and molecular studies have indicated that the extensive use or misuse of antibiotics in food animals for therapeutic and growth-promoting purposes can lead to the emergence of MDR bacteria (Kumar et al., 2019). In fact, frequent exposure of commensal bacteria of food animals to antimicrobial pressure can lead to the emergence of MDR bacteria, such as extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. This is of particular concern because those infected or asymptomatic animals can disseminate antimicrobial resistance into food chain and environment, leading to an increase in the spreading of resistant determinants via horizontal gene transfer among many bacterial species (Fluckey et al., 2007). Data obtained in the current study revealed that all Salmonella isolates, regardless of their origin displayed MDR phenotype, particularly to extended important antibiotics. The MIC values of all tested antibiotics were determined, and the results were interpreted according to the guidelines suggested by the Clinical and Laboratory Standards Institute (CLSI, 2015). In the current study, all Salmonella isolates displayed MIC values above the susceptibility breakpoint for ampicillin, piperacillin, and second-generation cephalosporins but showed reduced susceptibility to third-generation cephalosporins, suggesting that these isolates are potential ESBL producers. According to Laupland et al. (2010), the widespread application of third- and fourth-generation cephalosporins in food animals has resulted in a significant rise in bacteria that produce ESBL and AmpC β-lactamases, including Salmonella and Escherichia coli. These bacteria eventually serve as a reservoir of resistant bacteria for humans (Laupland et al., 2010). Such MDR phenotypes may spread quickly throughout hospitals and the community in Libya posing serious public health concerns (Ahmed et al., 2017; Zorgani et al., 2017) Some isolates in the current investigation showed an MIC above the fluoroquinolone (FQ) resistance breakpoint, which ranged from ≤0.25 to ≤4 µg/ml. A similar result was reported by (Abouzeed et al., 2008). Since more cases of FQ resistance have been documented, the WHO has designated FQ-resistant Salmonella spp. as a high prioritized pathogen (Cuypers et al., 2018). With the exception of one S. montevideo isolate that came from chickens and was susceptible to the majority of the antibiotics tested, all of the other isolates in this investigation were resistant to amikacin, tobramycin, and gentamicin. In conclusion, the current study demonstrated that high resistance rates to FQs can compromise important therapeutic options against salmonellosis. This also poses significant public health concerns. In this investigation, many isolates of Salmonella that were examined showed the MDR phenotype and high resistance rates against important antibiotic classes (such as ampicillin, cephalosporins, and FQs) that are frequently used to treat various infections in humans and animals. AcknowledgmentsNot applicable. Conflict of interestsThe authors declare that they have no competing interests. FundingThe author(s) received no financial support for the research, authorship, and/or publication of this article. Authors’ contributionsYMA, MOA, and SJA designed the plan of the manuscript, analyzed the data, and shared in writing and submitting the manuscript. HHA, JAA, SAF, and ESK validated the research experimental design and revised the manuscript. All authors read and approved the final manuscript. Ethics approvalNot applicable. Consent for publicationNot applicable. Data availabilityNot applicable. ReferencesAbouzeed, Y.M., Hariharan, H., Poppe, C. and Kibenge, F.S. 2000. Characterization of Salmonella isolates from beef cattle, broiler chickens, and human sources on Prince Edward Island. Comp. Immunol. Microbiol. Infect. Dis. 23(4), 253–266. Abouzeed, Y.M., Baucheron, S. and Cloeckaert, A. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52(7), 2428–2434. Ahmed, M.O., Aghila, E., Elamri, S. and Baptiste, K.E. 2017. Molecular investigation of carbapenemase-producing Enterobacteriaceae isolated from a Tripoli Hospital, Libya. Libyan J. Med. Sci. 1(3), 80–82. Altayyar, I.A., Elbreki, M.F., Ali, M.O. and Ali, A.A. 2016. Prevalence and antimicrobial susceptibility patterns of Salmonella spp. isolated from gastroenteritis patients, Southwestern, Libya. J. Appl. Med. Biomed. Res. 1(2), 2–6. Bouchrif, B., Paglietti, B., Murgia, M., Piana, A., Cohen, N., Ennaji, M.M., Rubino, S. and Timinouni, M. 2009. Prevalence and antibiotic-resistance of Salmonella isolated from food in Morocco. J. Infect. Dev. Ctries. 3(01), 035–040. Bouzidi, N., Aoun, L., Dekhil, M., Granier, S. A., Poirel, L., Brisabois, A., Nordmann, P. and Millemann, Y. 2011. Co-occurrence of aminoglycoside resistance gene armA in non-Typhi Salmonella isolates producing CTX-M-15 in Algeria. J. Antimicrob. Chemother. 66(9), 2180–2181. Centers for Disease Control and Prevention (CDC). 2010. Salmonella montevideo infections associated with salami products made with contaminated imported black and red pepper—United States, July 2009–April 2010. MMWR Morb. Mortal. Wkly. Rep. 59(50), 1647–1650. Chen, H.M., Wang, Y., Su, L.H. and Chiu, C.H. 2013. Nontyphoid Salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr. Neonatol. 54(3), 147–152. Clinical and Laboratory Standards Institute (CLSI). 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI Document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute. Cuypers, W.L., Jacobs, J., Wong, V., Klemm, E.J., Deborggraeve, S. and Van Puyvelde, S. 2018. Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb. Genom. 4(7), e000195. Elnageh, H.R., Hiblu, M.A., Abbassi, M.S., Abouzeed, Y.M. and Ahmed, M.O. 2021. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from cats and dogs in Tripoli, Libya. Vet. Ital. 57(2), 111–118. European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC). 2017. The European Union summary report on trends and sources of zoonoses, zoonotic agents, and food-borne outbreaks in 2016. EFSA J. 15(12), e05077. Fluckey, W.M., Loneragan, G.H., Warner, R. and Brashears, M.M. 2007. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J. Food Prot. 70(3), 551–556. Ghoddusi, A., Nayeri Fasaei, B., Zahraei Salehi, T. and Akbarein, H. 2019. Serotype distribution and antimicrobial resistance of Salmonella isolates in human, chicken, and cattle in Iran. Arch. Razi Inst. 74(3), 259–266. Glenn, L.M., Lindsey, R.L., Frank, J.F., Meinersmann, R.J., Englen, M.D., Fedorka-Cray, P.J. and Frye, J.G. 2011. Analysis of antimicrobial resistance genes detected in multidrug-resistant Salmonella enterica serovar Typhimurium isolated from food animals. Microb. Drug Resist. 17(3), 407–418. Gutema, F.D., Agga, G.E., Abdi, R.D., De Zutter, L., Duchateau, L. and Gabriël, S. 2019. Prevalence and serotype diversity of Salmonella in apparently healthy cattle: systematic review and meta-analysis of published studies, 2000–2017. Front. Vet. Sci. 6, 102. Kumar, D., Pornsukarom, S. and Thakur, S. 2019. Antibiotic usage in poultry production and antimicrobial-resistant Salmonella in poultry. In Food safety in poultry meat production. Eds., Venkitanarayanan, K., Thakur, S. and Ricke, S. Cham, Switzerland: Springer, pp: 47–66. Kurtz, J.R., Goggins, J.A. and McLachlan, J.B. 2017. Salmonella infection: interplay between the bacteria and host immune system. Immunol. Lett. 190, 42–50. Laupland, K.B., Schønheyder, H.C., Kennedy, K.J., Lyytikäinen, O., Valiquette, L., Galbraith, J., Collignon, P. and International Bacteremia Surveillance Collaborative. 2010. Salmonella enterica bacteraemia: a multi-national population-based cohort study. BMC Infect. Dis. 10, 95. Mshelbwala, F.M., Ibrahim, N.D.G., Saidu, S.N., Azeez, A.A., Akinduti, P.A., Kwanashie, C.N., Fakilahyel Kadiri, A.K., Muhammed, M., Fagbamila, I.O. and Luka, P.D. 2017. Motile Salmonella serotypes causing high mortality in poultry farms in three South-Western states of Nigeria. Vet. Rec. Open 4(1), e000247. O’Hara, C.M. and Miller, J.M. 2003. Evaluation of the Vitek 2 ID-GNB assay for identification of members of the family Enterobacteriaceae and other nonenteric gram-negative bacilli and comparison with the Vitek GNI+ card. J. Clin. Microbiol. 41(5), 2096–2101. Seiffert, S.N., Perreten, V., Johannes, S., Droz, S., Bodmer, T. and Endimiani, A. 2014. OXA-48 carbapenemase-producing Salmonella enterica serovar Kentucky isolate of sequence type 198 in a patient transferred from Libya to Switzerland. Antimicrob. Agents Chemother. 58(4), 2446–2449. World Health Organization (WHO). 2020. Salmonella (non-typhoidal). Available via https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) Zorgani, A., Almagatef, A., Sufya, N., Bashein, A. and Tubbal, A. 2017. Detection of CTX-M-15 among uropathogenic Escherichia coli isolated from five major hospitals in Tripoli, Libya. Oman Med. J. 32(4), 322. | ||
| How to Cite this Article |
| Pubmed Style Aishbani SJ, Al-griw HH, Sherif JA, Karim ES, Farag SA, Ahmed MO, Abouzeed YM. Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya. Open Vet. J.. 2025; 15(5): 1941-1946. doi:10.5455/OVJ.2025.v15.i5.8 Web Style Aishbani SJ, Al-griw HH, Sherif JA, Karim ES, Farag SA, Ahmed MO, Abouzeed YM. Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya. https://www.openveterinaryjournal.com/?mno=214237 [Access: January 24, 2026]. doi:10.5455/OVJ.2025.v15.i5.8 AMA (American Medical Association) Style Aishbani SJ, Al-griw HH, Sherif JA, Karim ES, Farag SA, Ahmed MO, Abouzeed YM. Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya. Open Vet. J.. 2025; 15(5): 1941-1946. doi:10.5455/OVJ.2025.v15.i5.8 Vancouver/ICMJE Style Aishbani SJ, Al-griw HH, Sherif JA, Karim ES, Farag SA, Ahmed MO, Abouzeed YM. Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya. Open Vet. J.. (2025), [cited January 24, 2026]; 15(5): 1941-1946. doi:10.5455/OVJ.2025.v15.i5.8 Harvard Style Aishbani, S. J., Al-griw, . H. H., Sherif, . J. A., Karim, . E. S., Farag, . S. A., Ahmed, . M. O. & Abouzeed, . Y. M. (2025) Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya. Open Vet. J., 15 (5), 1941-1946. doi:10.5455/OVJ.2025.v15.i5.8 Turabian Style Aishbani, Sumayah J., Huda H. Al-griw, Jihan A. Sherif, Elfurgani S. Karim, Samira A. Farag, Mohamed O. Ahmed, and Yousef M. Abouzeed. 2025. Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya. Open Veterinary Journal, 15 (5), 1941-1946. doi:10.5455/OVJ.2025.v15.i5.8 Chicago Style Aishbani, Sumayah J., Huda H. Al-griw, Jihan A. Sherif, Elfurgani S. Karim, Samira A. Farag, Mohamed O. Ahmed, and Yousef M. Abouzeed. "Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya." Open Veterinary Journal 15 (2025), 1941-1946. doi:10.5455/OVJ.2025.v15.i5.8 MLA (The Modern Language Association) Style Aishbani, Sumayah J., Huda H. Al-griw, Jihan A. Sherif, Elfurgani S. Karim, Samira A. Farag, Mohamed O. Ahmed, and Yousef M. Abouzeed. "Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya." Open Veterinary Journal 15.5 (2025), 1941-1946. Print. doi:10.5455/OVJ.2025.v15.i5.8 APA (American Psychological Association) Style Aishbani, S. J., Al-griw, . H. H., Sherif, . J. A., Karim, . E. S., Farag, . S. A., Ahmed, . M. O. & Abouzeed, . Y. M. (2025) Antibiotic susceptibility of Salmonella spp isolated from farm animals and environmental sources in Tripoli, Libya. Open Veterinary Journal, 15 (5), 1941-1946. doi:10.5455/OVJ.2025.v15.i5.8 |