| Research Article | ||
Open Vet J. 2025; 15(1): 252-260 Open Veterinary Journal, (2025), Vol. 15(1): 252-260 Research Article Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene)Siswani Siswani1,2*, Mochammad Hatta1, Muflihanah Muflihanah2, Nurul Muslinah Muhiddin1, Fitrine Ekawasti3, Rini Damayanti3, Riza Zainuddin Ahmad3, Putri Reno Intan4, Fitriana Fitriana4, Muhammad Ibrahim Desem4 and Ratih Rinendyaputri41Department of Microbiology, Faculty of Medicine, Hasanuddin University, Makassar, Indonesia 2Animal Diseases Investigation Maros, Directorate of Animal Health, Ministry of Agriculture, Maros, Indonesia 3Research Center for Veterinary Sciences, Research Organization for Health, National Research and Innovation Agency, Jakarta, Indonesia 4Research Center for Biomedis, Research Organization for Health, National Research and Innovation Agency, Jakarta, Indonesia *Corresponding Author: Siswani Siswani. Department of Microbiology, Faculty of Medicine, Hasanuddin University, Makassar, Indonesia and Animal Diseases Investigation Maros, Directorate of Animal Health, Ministry of Agriculture, Maros, Indonesia. Email: ssiswani83 [at] gmail.com Submitted: 11/09/2024 Accepted: 09/12/2024 Published: 31/01/2025 © 2025 Open Veterinary Journal
AbstractBackground: Indonesia is currently experiencing a foot-and-mouth disease (FMD) outbreak in livestock, caused by the FMD virus (FMDV). FMDV is easily spread to cause outbreaks in new geographical locations. Identifying FMDV infection through early diagnostic testing is important to track and prevent the spread of FMD in Indonesia. The use of appropriate samples is a crucial step in a study or survey to provide accurate results for diagnosis. Aim: The aim of our study was to evaluate the sensitivity of relevant oral mucosal epithelial swabs, hypersalivation swabs, and blood plasma specimen samples for FMDV diagnosis using one-step real time PCR (3D). Methods: Sampling was carried out by judgment or purposive, namely selecting animals that showed symptoms of the disease. A total of 109 samples were collected consisting of 3 types of oral mucosal epithelial swab specimens, hypersalivation swabs, and blood plasma collected from each cow reported as suspected FMDs in 8 districts in South Sulawesi were tested for FMDV using one-step RT-PCR (3D). Results: FMDV infection in oral mucosal epithelial swabs (ct=28.9), saliva (ct=30.62), and blood plasma (ct=37.17) compared to controls used from the Farma Veterinary Center (ct=29.10) using RT PCR (3D gene). These results indicate that the sensitivity value of oral mucosal epithelial swab specimens is the same as positive controls, even more sensitive, than when compared to saliva or blood plasma specimens. Sensitivity depends on the cycle threshold (CT) value for each RT PCR test and should be preferred in crucial situations if possible. Conclusion: These findings indicate that using oral mucosal epithelial swab specimens is more sensitive to be used as superior samples for FMDV detection using RT-PCR (3D) as a powerful tool for early detection to enable faster and effective treatment. Keywords: FMDV, RT-PCR, South Sulawesi, Specimen. IntroductionFoot-and-mouth disease (FMD) is a highly infectious viral disease spread by animals with open fork legs that affects up to 70 species of cloven-hoofed mammals including cattle, sheep, goats, and pigs (Jamal and Belsham, 2013; Knight-Jones and Rushton, 2013; Sameer and Jarullah, 2014). It is one of the leading causes of economic and cow losses and affects up to 70 species of cloven-hoofed mammals including cattle, sheep, goats, and pigs. It is one of the most serious illnesses impacting the international trade of animals and their products, according to the Office of International des Epizooties (World Organization for Animal Health, 2009). The FMD virus (FMDV) is the cause of FMD, and it is a member of the family Picornaviridae and genus Aphthovirus. FMDV is a tiny, non-enveloped, positive-sense, single-stranded RNA virus with a genome of roughly 8,500 nucleotides that is encased in an icosahedral capsid made up of four structural proteins. The FMDV virus has seven immunologically diverse serotypes (O, A, C, Asia 1, Southern African Territories 1, 2, and 3), each of which has a variety of antigenically and epidemiologically distinct subtypes (Jamal et al., 2012; Knowles et al., 2012). Clinical signs of FMD include fever and the development of vesicles in and around the mouth, foot, and mammary glands. These vesicles frequently quickly rupture and develop into erosive lesions. The additional signs include excessive salivation, anorexia, sadness, lameness, and resistance to rising or moving. Chronic lameness, a temporary or permanent drop in milk output, weight loss, mastitis, and condition loss are all potential complications. Multifocal myocarditis typically only causes death in young animals (Diab et al., 2019). FMD is a strategically contagious animal disease and is still exotic in Indonesia, and evidence of five of the seven FMDV serotypes has been reported in the country by several research publications (Adjid, 2020; Zainuddin et al., 2023). Even though there are routine programs to control and eradicate FMD, sometimes this disease still appears in several regions in Indonesia. South Sulawesi is one of the areas that is a priority for developing beef cattle farming in Indonesia (Eldiana et al., 2015). As the spread of FMD disease and the control that has been carried out, it is necessary to know information about the current incidence of FMD in South Sulawesi. Due to the economic implications and the public health hazards of FMD, the disease has commanded international attention for its control and prevention, which depends on early diagnosis, vaccination, and strict quarantine measures in addition to good animal care (El-Rahman et al., 2020; Firman et al., 2022). To anticipate the emergence of this disease, a quick and accurate diagnostic test is required. Real time PCR (RT-PCR) is a diagnostic test that is recommended by the World Animal Health Organization (OIE) (EL-Shehawy et al., 2012; Olabode et al., 2014; Tesfaye et al., 2020). FMDV molecular identification (fast and accurate) to detect coding region 3D in the FMDV genome. This research aims to evaluate the sensitivity of relevant oral mucosal epithelial swabs, hypersalivation swabs, and blood plasma specimen samples for FMDV diagnosis using one-step RT-PCR (3D). RT-PCR could be easily deployed for routine surveillance hence improving disease control measures. Materials and MethodsStudy period and locationSamples were collected from eight districts in South Sulawesi Province, Indonesia, throughout 2023. This location was selected because it had the highest cattle population in South Sulawesi. The location of these areas is shown in Figure 1. The average annual temperature is 26°C. Sample collectionA total of 109 samples were collected purposefully from cattle exhibiting clinical FMD signs, including cows with lesions in the mouth, gums, or foot and hypersalivation. The samples taken from each cow consisted of three types of specimens, namely oral mucosal swabs, saliva, and blood plasma (Fig. 2) and then placed into viral transport media (VTM). Research sample sizeDisease detection samples are usually used to detect the presence of disease in the population being investigated. The calculation of disease detection samples requires estimates of sampling confidence levels, disease prevalence, and the number of animals in the population. The disease detection sample formula is as follows(Martin et al., 1987): n=[1−(1−a)1/D] [N−(D1)/2], where the prevalence of the disease is estimated to be 5%, the sampling confidence level (a)=95%, so that the number of samples that must be taken is obtained, namely as many as 57 individuals (minimum sample). The method of sampling to detect disease is by judgment or purposive, namely selecting animals that show symptoms of disease. Sample preparationSpecimens include oral mucosal epithelial swabs, hypersalivation swabs, and blood plasma as well as other samples originating from animals suspected of being infected with FMD. The specimen was homogenized and added with VTM and then centrifuged at 4,000 G for 10 minutes to obtain the supernatant as a sample for RNA extraction. The positive control used isolates from the Pharma Veterinary Center Pusvetma, Directorate General of Animal Husbandry and Animal Health, Ministry of Agriculture. RNA extractionThe extraction was performed using the QiAmp Viral RNA Mini Kit (Qiagen) procedure. A total of 140 μl sample was added with 560 μl of a viral lysis buffer (AVL) buffer. The mixture was then homogenized using vortex and spindown. Furthermore, incubation was carried out for 10 minutes at room temperature. Centrifuge for 1 minute at 8,000 rpm (6,000 g) at 4°C. Add 560 μl of absolute ethanol and vortex. Centrifuge for 1 minute at 8,000 rpm (6,000 g) at 4°C. Transfer 630 μl of the solution into the Qiamp Spin Column. Centrifuge for 1 minute at 8,000 rpm (6,000 g) at 4°C. Discard all liquid in the collection tube. Transfer the remaining solution into the Qiamp Spin Column. Centrifuge for 1 minute at 8,000 rpm (6,000 g) at 4°C. Discard all liquid in the collection tube. Add 500 μl Buffer AW1. Centrifuge for 1 minute at 8,000 rpm (6,000 g) at 4°C. Discard all liquid in the collection tube. Replace the collection tube with a new one. Add 500 μl Buffer AW2, then centrifuge for 3 minutes at 14,000 rpm (20,000 g) at 4°C. Discard all liquid in the collection tube, then centrifuge for 1 minute at 8,000 rpm (6000 g) at 4°C. Transfer the Qiamp spin column to a new collection tube (1.5 ml), then add 50 μl buffer AVE and incubate for 1 minute. Centrifuge for 1 minute at 8,000 rpm (6,000 g) at 4°C. Discard the Qiamp spin column and label the tube. RNA products should be stored in a freezer at −20oC or −80oC. The commercial extraction kits were used according to the manufacturer’s instructions.
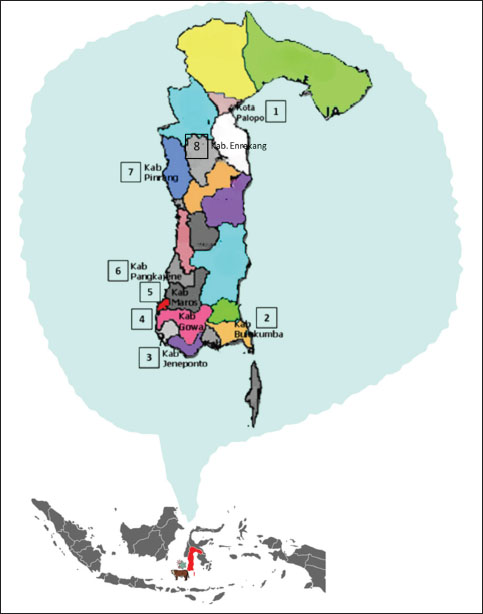
Fig. 1. Maps showing the Sulawesi islands in Indonesia and the locations of examined regency (Sumber: makassar.bpk.go.id).
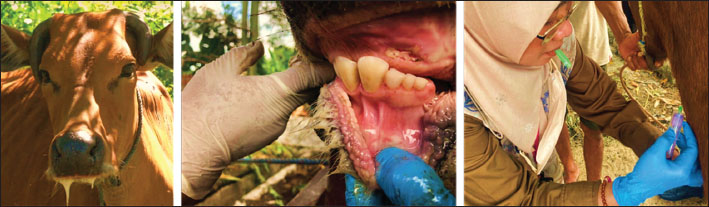
Fig. 2. Sampling of oral mucosa, saliva, and blood plasma specimens from cows showing clinical symptoms of FMD affected cattle (Animal Diseases Investigation Maros documentation). Real-time PCR FMDV 3DThe t-RNA amplification process for extracted specimens used the AgPath-ID One-step RT-PCR Kit (Ambion-ABI P/N AM1005) (manual-based procedure) and 1 µl of each primer. The primers/probe used in RT-PCR PMK testing 3D are Primer 3D 6769 F (5′-ACT GGG TTT TAC AAA CCT GTG A-3′); Primer 3D 6875 R (5′-GCG AGT CCT GCC ACG GA-3′); and 3D Primer 6820 P (5′-TCCTTTGCACGCCGTGGGAC′-BHQ1-3′). The FMDV 3D rRT-PCR was a modified version of the assay described by Callahan et al. (2002). The thermal cycling parameters for DNA amplification on the settings in the Applied Biosystems® 7,500 real-time PCR machine with thermal cycling conditions are as follows: (1 ×) 50°C for 2 minutes, 95°C for 10 minutes, (50 ×) 95°C for 15 seconds, and 60°C for 60 seconds. A positive control FMDV sample was included in each run as well as a negative water sample. The cutoff of the assay was set at a Ct-value of 45. Data analysisThe data obtained were analyzed descriptively with Microsoft Excel and using the analysis of variance (ANOVA) statistical program on SPSS version 17.0 statistical software package (SPSS Inc., Chicago, IL) using one-way between subjects. Differences were considered statistically significant at p-value of less than 0.05 (p value <0.05). Ethical approvalThis study was approved by Hasanuddin University Research Ethics Committee, University Faculty of Medicine (Approval No. 346/UN4.6-4.5.3L/ PP36 / 2023). ResultsClinically positive 109 samples with different kinds of specimens (oral mucosal epithelial swabs, saliva, and blood plasma) were evaluated by RT-PCR for the diagnosis of FMD using specific primers 3D. Samples producing a curve above the threshold value (ct) were considered positive. A graphic quantification of amplification showing the result, and melting curve on the PCR product is shown in Figure 3. The significant advantages of qPCR include its ability to measure DNA concentrations over a large range, sensitivity, ability to process multiple samples simultaneously, and ability to provide immediate information (Ekawasti et al., 2024). Unlike conventional PCR, real-time PCR uses fluorescence so that every time amplification occurs, a fluorescence signal will be formed, which will be captured by the detector throughout the PCR process. The amplification process is repeated in several cycles, and the resulting fluorescence signal will be directly proportional to the amplification that occurs. At one point, the number of fluorescence signals in the amplification process reaches a minimum value to be interpreted as a positive result. This point is called the cycle threshold (CT) value (Callahan et al., 2002). The amplification program settings, namely using passive reference, are selected NONE, reporter dye fluorescein amidine (FAM), and quencher NONE. The interpretation of the amplification results is seen from the CT value, namely if <40, then it is declared positive; between 40 and 45 is considered intermediate, and ≥45 is negative, and the PCR machine lists undetermined.
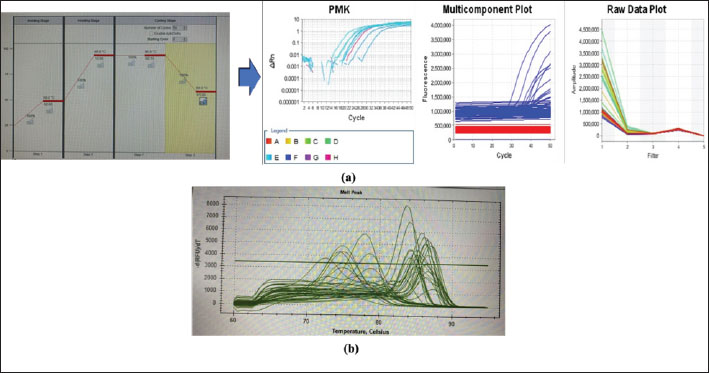
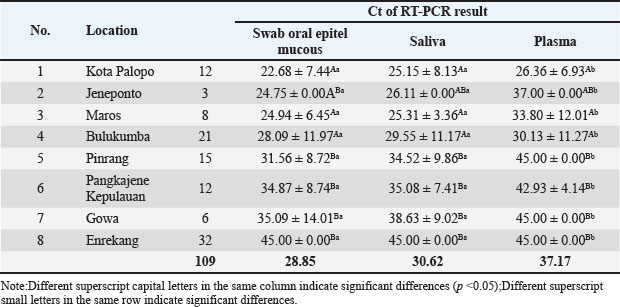
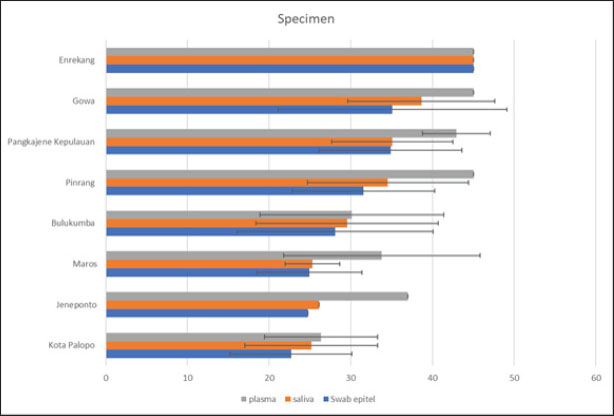
Fig. 3. (a) Real-time RT-PCR amplification by serotype-specific primers and probes 3D gene (b) Melting curve analysis optimization reaction. Table 1. Specimens sample in RT-PCR.
Fig. 4. Mean value of Ct result from specimen sample using RT-PCR (3D). Our results showed that the mean Ct value of samples from 8 districts in South Sulawesi Province were 28.90; 30.62; and 37.17 of FMD PCR positive for the oral mucosal epithelial swabs, saliva, and blood plasma specimens, respectively. Almost similar and sensitive results observed compared to positive controls from the Farma Veterinary Center used (ct=29.10) were oral mucosal epithelial swabs. These results indicate that the sensitivity value of oral mucosal epithelial swab specimens is the same as positive controls, even more sensitive, and also compared to saliva or blood plasma specimens. The ANOVA analysis showed significance (p > 0.05) between specimens (Table 1; Fig. 4). DiscussionThe virus called FMD is highly contagious. It is also a serious transboundary animal infection. Because of its severe impact on trade and manufacturing, FMD can have crippling socioeconomic impacts (Alexanderson, 2002; Callahan et al., 2002). The livestock industry suffers significant losses from FMD due to the high costs of prevention or control methods, such as mass vaccination campaigns and/or culling of infected animals, as well as the clinical impact of the disease on milk and beef production. Vesicular lesions, associated with fever, lameness, drooling, and anorexia in clinically ill cattle, can be found on the mouth, feet, and udder (Paixão et al., 2008; Directorate General of Livestock and Animal Health Service, 2022; Sutawi et al., 2023). The cattle farming sector in South Sulawesi continues to grow, even sending superior cattle to many other provinces in Indonesia. South Sulawesi has been designated as a national cattle breeding center by the Ministry of Agriculture. This decision was not made lightly; many factors must be considered, including a supportive agro-climate and the farming traditions of the population, which have been passed down from generation to generation (Ministry of Agriculture of the Republic of Indonesia, 2022). It is critical to do an early study and identify this disease using the appropriate detection technology in order to implement a speedy control approach. The correct identification procedure is employing the appropriate sample and method to produce accurate results. Cattle in South Sulawesi exhibiting FMD symptoms, as reported by farmers to the Maros Veterinary Center, were sampled using a sampling technique aiming at clinical symptoms of FMD in cattle. FMDV-infected animals typically develop lesions on the tongue, snout, oral cavity, coronary band, and teats. Other typical symptoms include fever, poor appetite, weight loss, hypersalivation, depression, growth retardation, and considerably decreased milk production, which may last even after recovery (OIE, 2018). On the other hand, diagnosis based on clinical symptoms is problematic because many diseases have similar symptoms; therefore, laboratory confirmation is required for each suspected FMD patient (Wong et al., 2020). Several approaches can be used to confirm the presence of the FMDV, including RT-PCR, which is an accurate and sensitive FMD detection tool. The most appropriate sample usage may differ based on the type of target virus to be discovered and the characteristics of the person being tested. The FMD samples include oral mucosal swabs, saliva, and blood plasma (Wong et al., 2020). Sampling was carried out at several locations in South Sulawesi with different geographical areas (Fig. 1). The increase in sensitivity in determining the diagnosis was reflected in the increase in the detection ability of sample types and the level of serotype determination of clinical samples from FMD outbreaks in the field (Lim et al., 2022). Comparison of the RT-PCR sensitivity across three types of FMD sample specimens from the same cattle using RT-PCR. The 3D RT-PCR method is a method that has been validated by FAO, so researchers only compare the results of the types of sample specimens used. In this study, RT-PCR was carried out on 109 clinical samples. To detect FMDV RNA, this primer probe is used in several international studies (Jamal and Belsham, 2013; Reid et al., 2014; Jamal and Belsham, 2015). Based on the table, using a 5% level of significance, it can be interpreted as follows: There is no significant difference between oral mucosal epithelial swabs and saliva. There is a significant difference between oral mucosal epithelial swabs and plasma; there is a significant difference between saliva and plasma. Furthermore, categorization is carried out for the average value of each specimen and location, showing that with an average interval value of Ct value ≤ 22.5, 22.6–44.9, and ≥ 45 indicates that the results are at high positive, low positive, and negative levels for FMDV, respectively. Table 1 reveals that Palopo City has the most sensitive oral mucosal swab specimen samples and the highest positive FMD incidence rate, while Enrekang shows negative results for three sample specimens, despite the presence of clinical symptoms in the cows sampled. This is in accordance with the statement by Lim et al. (2022) that clinical symptoms of FMD in cattle at the location showed symptoms of FMD but needed to be supported by laboratory tests so that FMD could be diagnosed accurately. DNA can also be identified from oral mucosal samples, although these samples are frequently harder to acquire than saliva and call for exacting methods. However, in cases where saliva samples are inadequate or tainted, this can be helpful. The sensitivity of FMD DNA detection in saliva and oral mucosal swab samples did not differ statistically significantly from one another, as indicated by the ANOVA p-value analysis results. This is in contrast to plasma samples, which are significantly different from saliva samples. Plasma is more frequently employed in chemical analysis since it is a liquid component of blood that contains a variety of chemicals. The results of this study strengthen the statement of Estevez Garcia et al., (2022) that when identifying respiratory viruses, such as FMD, oral mucosal swabs and saliva are frequently more sensitive than plasma. Saliva and oral mucosal swabs are sites where FMDV adhere or proliferate. The mouth and throat are two areas of the upper respiratory tract where respiratory viruses, including FMD, frequently proliferate and disseminate, making them possible infection sites. If the virus is present, even in trace amounts, this can improve the likelihood of finding it. Saliva and oral mucosal swabs serve to limit the possibility of contamination in plasma samples, which can lower the test’s specificity (Poonsuk et al., 2018). Saliva collection is one example of a noninvasive sampling technique that may be a more affordable option or supplement to current survey techniques (Garner et al., 2021). Saliva samples with low viral loads can still be identified using suitable enrichment techniques, suggesting that DNA-based molecular surveillance may be used (Mathijs et al., 2016). In the case of animal diseases such as FMD, it is crucial to remember that prompt and precise detection is necessary to stop the disease’s spread. In order to monitor and control this disease in animals, it is crucial to use the most suitable and sensitive sample technique. Furthermore, since the production of copious amounts of saliva is one of the clinical indications of FMD in cattle, it is possible to identify saliva that contains high levels of FMDV based on the results of the analysis of the huge amounts of DNA present in saliva. One of the potential ways that viruses might spread from animals to farms is through contaminated farm floors and contaminated farm equipment, such as animal feed, footwear, and equipment (Bravo de Rueda et al., 2014). In the meantime, the study’s findings give a general overview of the 3D gene RT-PCR technique, particularly the kind of sample specimen that is used to produce precise FMD diagnosis results. Preventing the spread of FMD requires early, accurate diagnosis and, of course, biosecurity precautions. ConclusionIn summary, this research supports the idea that oral mucosal swabs and saliva samples can be used in the RT-PCR test. Oral mucosal epithelial swab specimens are samples that have high sensitivity for using one-step real-time polymerase chain reaction (3D gene) test. This testing is to support early control decisions during disease outbreaks. Faster and better diagnostic methods can significantly improve control in both free and affected areas. AcknowledgmentsThe authors express sincere gratitude to the Faculty of Medicine, Hasanuddin University and Animal Diseases Investigation Maros, Directorate of Animal Health, Ministry of Agriculture. Conflict of interestThe authors declare that there is no conflict of interest. FundingThis research received no specific grant. Author’s contributionsSS, MH, MM, FE: Study conception and design. SS, MM, NMM, FE, RD: Conducted the experiments and analyzed the data. SH, FE, RZA, FF, PRI: Contributed to writing and revising the manuscript. SS, MID, FF, RR: participated in the preparation of the manuscript. All authors have read, reviewed, and approved the final manuscript. Data availabilityAll data are available in the manuscript. ReferencesAdjid, R.M. 2020. Foot and mouth disease: exotic animal disease that must be alert of entry into Indonesia. Wartazoa Vet. Sci. 30(2), 61–70. Alexanderson, S., Zhang, Z. and Donaldson, A.I. 2002. Aspects of the persistence of foot and mouth disease virus in animals-the carrier problem. Microbes Infect. 4(10), 1099–1110. Bravo de Rueda, C., Dekker, A., Eblé, P.L. and de Jong, M.C. 2014. Identification of factors associated with increased excretion of foot-and-mouth disease virus. Prev. Vet. Med. 113(1), 23–33. Callahan, J.D., Brown, F., Osorio, F.A., Sur, J.H., Kramer, E., Long, G.W., Lubroth, J., Ellis, S.J., Shoulars, K.S., Gaffney, K.L., Rock, D.L. and Nelson, W.M. 2002. Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J. Am. Vet. Med. Assoc. 220(11), 1636–1642. Directorate General of Livestock and Animal Health Service (DGLAHS) and Ministry of Agriculture of the Republic of Indonesia. 2022 Spread of FMD cases. Available via https://siagapmk.crisis-center.id/ (Accessed 1 September 2024). Diab, E., Bazid, A.I., Fawzy, M., El-Ashmawy, W.R., Fayed, A.A. and El-Sayed, M.M. 2019. Foot and mouth disease outbreaks in Egypt during 2013–2014: molecular characterization of serotypes A, O, and SAT2. Vet. World. 12(2), 190–197. Ekawasti, F., Winarsongko, A., Nepho, F., Purwanto, E.S., Subekti, D.T., Nuradji, H., Dharmayanti, N.L.P.I., Ahmad, R.Z., Sa’diah, S., Cahyaningsih, U. and Nurcahyo, R.W. 2024. Optimization of sybr green quantitative real time polymerase chain reaction (qPCR) using excreted-secreted antigens (ESAs) genetik marker for detection Toxoplasma gondii. Jurnal Sain. Vet. 42(1), 1–13. Eldiana, R., Baba, S. and Abdullah, A. 2015. The role of social capital in Beef Cattle Marketing Institutions in Bantimurung District, Maros regency. JIIP Natl. J. 2(2), 82–94. El-Rahman, M.M.A., El-Hassan, D.G.A., Awad, W.S. and Salem, S.A. 2020. Serological evaluation for the current epidemic situation of foot and mouth disease among cattle and buffaloes in Egypt. Vet. World. 13(1), 1–9. EL-Shehawy, L., Azab, A.M.H., Mossad, W., El-Sayed, E., Ismail, A. and Deghady, W. 2012. Real time RT-PCR assay for detection of different serotypes of FMDV in Egypt, Vet. World. 5(12), 732–737. Estevez Garcia, A.I., Lefebvre, D.J., Nyabongo, L., Haegeman, A., Nkundwanayo, C., De Vleeschauwer, A., Ntakirutimana, D., De Leeuw, I., Nsanganiyumwami, D., Niyokwizera, P., van den Berg, T., Niyokwishimira, A. and Clercq, K. 2022. Outbreaks of foot-and-mouth disease in Burundi, East Africa, in 2016, caused by different serotypes. Viruses 14(5), 1077. Firman, A., Trisman, I. and Puradireja, R.H. 2022. Economic impact of foot and mouth diseases outbreak on cattle and buffalo in Indonesia. Mimbar Agrib. 8(2), 1123–1129. Garner, G., Vosloo, W., Tapsuwan, S., Bradhurst, R., Seitzinger, A. H., Breed, A.C. and Capon, T. 2021. Comparing surveillance approaches to support regaining free status after a foot-and-mouth disease outbreak. Prev. Vet. Med. 194(105441), 1–12. Jamal, S.M. and Belsham, G.J. 2013. Foot-and-mouth disease: past, present and future. Vet. Res. 44(116), 1–14. Jamal, S. M., and Belsham, G. J. 2015. Development and characterization of probe-based real time quantitative RT-PCR assays for detection and serotyping of foot-and-mouth disease viruses circulating in West Eurasia. PloS One. 10(8), 1–16. Jamal, S.M., Ferrari, G., Hussain, M., Nawroz, A.H., Aslami, A.A., Khan, E., Murvatulloev, S., Ahmed, S. and Belsham, G.J. 2012. Detection and genetic characterization of foot-and-mouth disease viruses in samples from clinically healthy animals in endemic settings. Transbound. Emerg. Dis. 59(5), 429–440. Knight-Jones, T.J.D. and Rushton, J. 2013. The economic impacts of foot and mouth disease-what are they, how big are they and where do they occur? Prev. Vet. Med. 112(3–4), 161–173. Knowles, N.J., Hovi, T., Hyypiä, T., King, A.M.Q., Lindberg, A.M., Pallansch, M.A, Palmenberg, A.C., Simmonds, P., Skern, T., Stanway, G., Yamashita, T. and Zell, R. 2012. Picornaviridae. In Virus taxonomy: classification and nomenclature of viruses. 9th report of the international committee on taxonomy of viruses. Eds., King, A.M.Q., Adams, M.J., Carstens, E.B. and Lefkowitz, E.J. San Diego, CA: Elsevier, pp: 855–880. Lim, D.R., Ryoo, S., Kang, H., Oh, S.H., Jang, S.H., Kang, B., Park, H.J., Hwang, H., Kim, J.M., Park, C.K. and Cha, S.H. 2022. Enhanced detection and serotyping of foot-and-mouth disease virus serotype O, A, and Asia1 using a novel multiplex real-time RT-PCR. Transbound Emerg. Dis. 69(5), e2578–e2589. Martin, S., Meek, A. and Preben, W. 1987. Veterinary epidemiology—principles and methods, 1st ed. Ames, IA: Iowa State University Press. Mathijs, E., Vandenbussche, F. and Van Borm, S. 2016. Using genomics for surveillance of veterinary infectious agents. Rev. Sci. Tech. 35(1), 143–157. Ministry of Agriculture of the Republic of Indonesia (MoARI) and Directorate of Animal Health, Animal Diseases Investigation Maros. 2022. Control and management of mouth and hook diseases in animals (FMD) Center. Available via https://bbvet-maros-ppid.pertanian.go.id/index.php/news/view/1589 (Accessed 1 September 2024). OIE. 2018. Foot & mouth disease (FMD). Available online at: http://www.oie.int/en/animal-health-in-the-world/animal-diseases/Footand-mouth-disease/ (Accessed 1 September 2024) Olabode, O.H., Kazeem, H.M. and Raji, M.A. 2014. Diagnosis of bovine foot and mouth disease virus by real-time polymerase chain reaction and nucleotide sequencing from outbreak herd samples in Ilesha Baruba, Kwara state, Nigeria, Vet. World. 7(10), 868–875. Paixão, T.A., Neta, A.V., Paiva, N.O., Reis, J.R., Barbosa, M.S., Serra, C.V., Silva, R.R., Beckham, T.R., Martin, B.M., Clarke, N.P., Adams, L.G. and Santos, R.L. 2008. Diagnosis of foot-and mouth disease by real time reverse transcription polymerase chain reaction under field conditions in Brazil. BMC Vet. Res. 31(4), 1–6. Poonsuk, K., Giménez-Lirola, L. and Zimmerman, J. 2018. A review of foot-and-mouth disease virus (FMDV) testing in livestock with an emphasis on the use of alternative diagnostic specimens. Anim. Health Res. Rev. 19(2), 100–112. Reid, S.M., Grierson, S.S., Ferris, N.P., Hutchings, G.H. and Alexandersen, S. 2003. Evaluation of automated RTPCR to accelerate the laboratory diagnosis of foot-and-mouth disease virus. J. Virol. Methods. 107(2), 129–139. Sameer, M.A. and Jarullah, B.A. 2014. Epidemiological study of foot and mouth disease and evaluation of vaccination method for controlling disease in Waset province. J. Univ. Thi-Qar, 9(2), 1–6. Sutawi, S., Wahyudi, A., Malik, A., Suyatno, S., Hidayati, A., Rahayu, I.D. and Hartatie, E.S. 2023. Re-emergence of foot and mouth disease outbreak in Indonesia: a review. Adv. Anim. Vet. Sci. 11(2), 264–271. Tesfaye, Y., Khan, F. and Gelaye, E. 2020. Molecular characterization of foot-and-mouth disease viruses collected from Northern and Central Ethiopia during the 2018 outbreak. Vet. World. 13(3), 542–548. Wong, C.L., Yong, C.Y., Ong, H.K., Ho, K.L. and Tan, W.S. 2020. Advances in the diagnosis of foot-and-mouth disease. Front. Vet. Sci. 7(477), 1–24. World Organization for Animal Health. 2009. Manual of diagnostic tests and vaccines for terrestrial animals. Paris, France: World Organisation for Animal Health, Office International Des Epizooties, pp:1–25. Zainuddin, N., Susila, E.B., Wibawa, H., Daulay, R.S.D., Wijayanti, P.E., Fitriani, D., Hidayati, D.N., Idris, S., Wadsworth, J., Polo, N., Hicks, H.M., Mioulet, V., Knowles, N.J. and King, D.P. 2023. Genome sequence of a foot-and-mouth disease virus detected in Indonesia in 2022. Microbiol. Resour. Announc. 12(2), e0108122. | ||
| How to Cite this Article |
| Pubmed Style Siswani S, Hatta M, Muflihanah M, Muhiddin NM, Ekawasti F, Damayanti R, Ahmad RZ, Intan PR, Fitriana F, Desem MI, Rinendyaputri R. Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene). Open Vet J. 2025; 15(1): 252-260. doi:10.5455/OVJ.2025.v15.i1.23 Web Style Siswani S, Hatta M, Muflihanah M, Muhiddin NM, Ekawasti F, Damayanti R, Ahmad RZ, Intan PR, Fitriana F, Desem MI, Rinendyaputri R. Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene). https://www.openveterinaryjournal.com/?mno=220004 [Access: July 15, 2025]. doi:10.5455/OVJ.2025.v15.i1.23 AMA (American Medical Association) Style Siswani S, Hatta M, Muflihanah M, Muhiddin NM, Ekawasti F, Damayanti R, Ahmad RZ, Intan PR, Fitriana F, Desem MI, Rinendyaputri R. Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene). Open Vet J. 2025; 15(1): 252-260. doi:10.5455/OVJ.2025.v15.i1.23 Vancouver/ICMJE Style Siswani S, Hatta M, Muflihanah M, Muhiddin NM, Ekawasti F, Damayanti R, Ahmad RZ, Intan PR, Fitriana F, Desem MI, Rinendyaputri R. Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene). Open Vet J. (2025), [cited July 15, 2025]; 15(1): 252-260. doi:10.5455/OVJ.2025.v15.i1.23 Harvard Style Siswani, S., Hatta, . M., Muflihanah, . M., Muhiddin, . N. M., Ekawasti, . F., Damayanti, . R., Ahmad, . R. Z., Intan, . P. R., Fitriana, . F., Desem, . M. I. & Rinendyaputri, . R. (2025) Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene). Open Vet J, 15 (1), 252-260. doi:10.5455/OVJ.2025.v15.i1.23 Turabian Style Siswani, Siswani, Mochammad Hatta, Muflihanah Muflihanah, Nurul Muslinah Muhiddin, Fitrine Ekawasti, Rini Damayanti, Riza Zainuddin Ahmad, Putri Reno Intan, Fitriana Fitriana, Muhammad Ibrahim Desem, and Ratih Rinendyaputri. 2025. Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene). Open Veterinary Journal, 15 (1), 252-260. doi:10.5455/OVJ.2025.v15.i1.23 Chicago Style Siswani, Siswani, Mochammad Hatta, Muflihanah Muflihanah, Nurul Muslinah Muhiddin, Fitrine Ekawasti, Rini Damayanti, Riza Zainuddin Ahmad, Putri Reno Intan, Fitriana Fitriana, Muhammad Ibrahim Desem, and Ratih Rinendyaputri. "Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene)." Open Veterinary Journal 15 (2025), 252-260. doi:10.5455/OVJ.2025.v15.i1.23 MLA (The Modern Language Association) Style Siswani, Siswani, Mochammad Hatta, Muflihanah Muflihanah, Nurul Muslinah Muhiddin, Fitrine Ekawasti, Rini Damayanti, Riza Zainuddin Ahmad, Putri Reno Intan, Fitriana Fitriana, Muhammad Ibrahim Desem, and Ratih Rinendyaputri. "Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene)." Open Veterinary Journal 15.1 (2025), 252-260. Print. doi:10.5455/OVJ.2025.v15.i1.23 APA (American Psychological Association) Style Siswani, S., Hatta, . M., Muflihanah, . M., Muhiddin, . N. M., Ekawasti, . F., Damayanti, . R., Ahmad, . R. Z., Intan, . P. R., Fitriana, . F., Desem, . M. I. & Rinendyaputri, . R. (2025) Sensitivity of specimen type for diagnosing foot-and-mouth diseases in cattle using one-step real-time polymerase chain reaction (3D gene). Open Veterinary Journal, 15 (1), 252-260. doi:10.5455/OVJ.2025.v15.i1.23 |