| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 459-469 Original Research Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin-resistant genes from poultry farms in EgyptReem I. Ali1, El-Sayed A. Abdelaziz1, Mohammed A. Kamel1, Susan K. Murad2, Hussam M. Abdallah3 and Gamal A. Salem1,4*1Department of Pharmacology, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt 2Department of Public Health, Faculty of Health Sciences, Misurata University, Misurata, Libya 3Department of Microbiology, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt 4Department of Medical Genetics, Faculty of Health Sciences, Misurata University, Misurata, Libya *Corresponding Author: Gamal A. Salem. Department of Pharmacology, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt. Email: gamal_vet_85 [at] yahoo.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/07/2023 © 2024 Open Veterinary Journal
AbstractBackground: Escherichia coli (E. coli) bacteria that produce extended spectrum beta-lactamase (ESBL) is associated with a high prevalence of human illnesses worldwide. The emergence of resistance to carbapenem and colistin compounds poses further challenges to the treatment options for these illnesses. This study aimed to evaluate the phenotypic and genotypic pattern of resistance to carbapenem and colistin in ESBL-producing E. coli. Escherichia coli isolates collected from the respiratory tract of chickens in El-Sharkia government, Egypt. Methods: A total of 250 lung samples were collected from 50 poultry farms. These samples were then subjected to isolation, identification, and serotyping of E. coli. The presence of antimicrobial resistance was identified by disc diffusion testing. The occurrence of ESBL phenotypes was also assessed using the double disc synergy method. PCR/sequencing techniques were employed to examine the presence of ESBL (β-lactamase (bla)-TEM, blaSHV, and blaCTX-M), colistin (mcr-1), and carbapenem (blaNDM, blaVIM, and blaKPC) resistance genes. Results: The findings revealed that 140 out of 250 (56%) were identified as E. coli. All E. coli isolates had a high level of multi-antimicrobial resistance (MAR) with an index value greater than 0.2, and 65.7% of them were confirmed to produce ESBL. Out of the 92 ESBL phenotypes, 55 (59.7%), 32 (34.7%), 18 (19.6%), and 37 (40.2%) isolates harbor b laTEM-3, b laSHV-4, b laCTX-M-1, a nd blaCTX-M-14 genes, respectively. The blaNDM-1 gene was identified in all 40 phenotypes that exhibited resistance to carbapenem, accounting for 28.5% of all strains of E. coli and 43.4% of ESBL isolates. The VIM and KPC genes were not detected in any of the samples. Furthermore, there was a significant prevalence of the mobilized colistin resistance (mcr)-1 gene, with 64 (69.5%) of the ESBL isolates exhibiting this gene. Conclusion: The prevalence of ESBL-producing E. coli, particularly those resistant to carbapenem and colistin, poses a significant public health risk in society. Keywords: Antibiotic resistance, Carbapenem, Colistin, E. coli, ESBL. IntroductionEscherichia coli, a bacterium belonging to the Enterobacteriaceae family, can cause infections in the intestines and other parts of the body in both people and animals. The emergence of extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae, which are resistant to third- and fourth-generation cephalosporins, has posed a challenge to the management of these infections since the 2000s (Poirel et al., 2018). There was evidence that E. coli originating from animals or animal-derived food possess zoonotic potential. During the last twenty years, there has been a significant increase in the prevalence of ESBL-producing E. coli which has presented a substantial risk to public health (Cardozo et al., 2021; Giufrè et al., 2021). The "One Health" strategy has been endorsed by organizations such as the World Organization for Animal Health, the Food and Agriculture Organization of the United Nations, and the World Health Organization in light of the global proliferation of antibiotic-resistant bacteria. This strategy entails collaborative endeavors among environmental, veterinary, and public health authorities to alleviate the dissemination of antibiotic resistance (White and Hughes, 2019). The transmission of resistant E. coli, which has ESBL-producing potential, is known to be influenced by direct contact between infected animals and humans. The sharing of resistance genes across animals and humans is attributed to sharing the same environment. The transmission of ESBL-producing E. coli from poultry or poultry products to humans has been recorded (Coppola et al., 2020). Based on the comparisons of amino acid sequences, ESBL variants can be classified into nine distinct structural families, which are blaTEM, blaSHV, blaCTX-M, blaPER, blaVEB, blaGES, blaBES, and blaTLA. The foremost ESBL variants are blaTEM, blaSHV, and blaCTX-M. The CTX-M variant, specifically, has rapidly disseminated and is widely documented in E. coli globally (Bubpamala et al., 2018). Carbapenems, such as ertapenem, meropenem, and imipenem, are a class of potent antibiotics that have a wide range of activity against many types of bacteria. They belong to the beta-lactam chemical group and are mostly employed in the treatment of severe infections in humans. These substances are well acknowledged for their exceptional efficacy and are frequently employed as a final option to combat bacteria that are resistant to many drugs, both Gram-negative and Gram-positive (Taggar et al., 2020). Nevertheless, they lack authorization for application in animal or veterinary domains. Consequently, the examination for carbapenem resistance is not commonly carried out in animals. Nevertheless, several extended-spectrum cephalosporins, such as cefquinome, ceftiofur, cefoperazone, and cefpodoxime, are commonly employed in the veterinary field. Ceftiofur, a third-generation cephalosporin, is frequently employed for the treatment of bacterial illnesses in livestock, including pneumonia, metritis, septicemia, and meningitis. Hence, the utilization of these antibiotics may apply selection pressure that facilitates the development of carbapenem-resistant bacteria. There is a higher occurrence of carbapenem-resistant Enterobacteriaceae (CRE), especially E. coli, Klebsiella pneumoniae, and Proteus mirabilis, carrying blaNDM genes in broiler farms. Among these genes, blaNDM-1 and blaNDM-5 are particularly prevalent (Liakopoulos et al., 2018). The primary cause of carbapenem resistance in Enterobacteriaceae is the synthesis of carbapenemases, which are frequently encoded by genes found on plasmids. Carbapenemases are categorized into three classes: Class A (KPC), Class B (NDM, IMP, VIM), and Class D (OXA-type) (Ambler, 1980). In impoverished countries such as Egypt, where there is often a lack of regulation on antimicrobial usage, colistin is commonly employed in veterinary practice (Moawad et al., 2017). Colistin, or polymyxin E, exhibits structural resemblances to polymyxin B. Although colistin demonstrates swift bactericidal activity against Gram-negative bacteria in laboratory conditions, its effectiveness is significantly diminished when used in living organisms. To overcome this constraint, the use of colistin can be augmented by incorporating other antibiotics into therapy regimens (Paul et al., 2018). The rise of colistin resistance is alarming in the field of human medicine since this medication is regarded as a final resort for treating serious infections caused by Enterobacteriaceae that are resistant to many drugs (Mendelson et al., 2018). Originally, chromosomal changes were thought to be the primary cause of colistin resistance. However, research conducted in multiple countries has revealed that resistance can also occur through alternative pathways. A recent study by Osei Sekyere (2019) has recorded the finding of nine mobilized colistin resistance (mcr) genes, namely mcr-1 to mcr-9. These genes are responsible for plasmid-mediated resistance against colistin. Global attention has been directed to the "one-health" idea and the potential danger posed by antibiotic-resistant germs in farm animals that can be passed to humans. There is a lack of comprehensive data on the prevalence of carbapenem and colistin-resistant Enterobacteriaceae in livestock that come into contact with the human community in Egypt. To investigate this matter, we undertook a study to determine the occurrence of carbapenem and colistin-resistant strains among ESBL-generating E. coli isolated from chicken farms in the eastern region of the Egyptian Delta. Furthermore, the study examined the genetic determinants of ESBL, carbapenem, and colistin resistance in E. coli within this region. Material and MethodsSample collectionThe collection of samples was conducted from 50 poultry farms situated in the eastern portions of the Egyptian delta, notably in El-Sharqia Governorate, spanning from December 2019 to April 2021. Five diseased chickens, ranging in age from 1 to 5 weeks and exhibiting signs of sickness, were obtained from each farm. The gathered chickens had signs including depression, reduced desire to eat, and disheveled plumage. The birds were conveyed to the Reference Laboratory for Veterinary Quality Control on Poultry Production, AHRI, Giza, and to the Faculty of Veterinary Medicine, Zagazig University, Egypt. The samples underwent post-mortem inspection under sterile conditions. Specimens were collected from the lung and trachea of birds exhibiting symptoms of colisepticemia, air sacculitis, pericarditis, and perihepatitis. The samples were dispatched for the purpose of bacterial isolation and identification. Isolation and identificationEscherichia coli strains were isolated and identified using the following procedure (Swayne, 1998). The samples were submerged in buffer peptone water and subjected to aerobic incubation for a duration of 24 hours at a temperature of 37°C. The samples were applied onto eosin methylene blue agar and MacConkey agar plates, and then placed in an incubator at a temperature of 37°C for a duration of 24 hours. The colonies suspected on MacConkey agar displayed a vibrant pink hue and measured 1–2 mm in diameter, but they exhibited a metallic color on eosin methylene blue agar. Presumed pure E. coli colonies underwent additional conventional biochemical screening, including tests for oxidase activity, citrate utilization, indole production, methyl red reaction, Voges Proskauer test, and triple sugar iron test. In addition, the process of serotyping the isolated E. coli strain is performed. The identification of E. coli strains was conducted using somatic (O) antigen antisera obtained from DENKA SEIKEN Company, Tokyo, Japan, following the instructions provided with the kit. Antimicrobial susceptibility testing (AST)The Mueller–Hinton agar (MHA) disc diffusion test is utilized to assess the antimicrobial susceptibility of all E. coli strains using a method previously outlined by Osei Sekyere (2019). The test comprised 12 antibiotic discs obtained from Oxoid (Basingstoke, UK), which included those utilized for the initial screening of ESBL, as well as carbapenems and colistin resistance. The screening process entailed conducting tests on the isolates using ceftazidime (30 µg), cefotaxime (30 µg), colistin (10 µg), and ertapenem (10 µg). The interpretation of inhibition zones was conducted in accordance with the guidelines set by the Clinical and Laboratory Standards Institute (CLSI). The double disc synergyTo verify the presence of ESBL, this test was conducted using a method previously described by Anago et al. (2015). The test involved applying amoxicillin-clavulanic acid (AMC, 30 µg) at a distance of 20 mm from each antibiotic disc (30 µg) containing third-generation cephalosporins (cefotaxime and ceftriaxone) and fourth-generation cephalosporins (ceftazidime and cefepime) on MHA plates. If the boundary of the area where the growth of bacteria is inhibited by cephalosporin extends significantly toward the AMC disc, it is considered a favorable indication of carrying ESBL (Fig. 1). Genotypic characterizations of E. coli isolatesGenotypic analysis was performed on all isolates that exhibited phenotypic ESBL production and multi-antimicrobial resistance (MAR). Cultured bacteria were subjected to DNA extraction using a Purification Kit (Roche Diagnostics, Mannheim, Germany) following the instructions provided by the manufacturer. In summary, a 200 μL sample of bacterial culture was combined with 200 μL of lysis buffer and 10 μL of proteinase K. The mixture was then subjected to incubation at a temperature of 56°C for a duration of 10 minutes. Afterward, the lysate was subjected to treatment with 100% ethanol (200 μL). Following washing and centrifugation, 100 μL of elution buffer was utilized to extract the nucleic acid. Every E. coli isolate underwent testing to determine the presence of resistance genes, including blaTEM, blaSHV, blaCTX-M, blaVIM, blaKPC, blaNDM-1, and mcr-1. Employing designated primers provided by Metabion (Planegg, Germany), as previously demonstrated in Table 1. The DNA that was obtained was used in PCR experiments using the Emerald Amp Max PCR Master Mix from Takara, Japan. The amplification was conducted on an Applied Biosystems 2,720 thermal cycler. The PCR protocol involved a single initial denaturation phase at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at a temperature range of 53°C–60°C for 30–40 seconds, and extension at 72°C for 30–45 seconds. Finally, there was a final extension cycle at 72°C for 7–10 minutes. The PCR products were separated using electrophoresis in 1× TBE buffer at room temperature. A 5 V/cm gradient was applied on a 1.5% agarose gel acquired from Applichem GmbH in Darmstadt, Germany. Each gel hole was filled with 20 µl of the products. The GelPilot 100 bp plus DNA Ladder, manufactured by Qiagen GmbH in Hilden, Germany, was employed to ascertain the sizes of the DNA fragments. The PCR results were subjected to purification and subsequently sequenced using the Sanger ABI 3730 XL automated DNA sequencer. The sequencing was performed by BaseClear, a commercial business located in Leiden, The Netherlands. The nucleotide sequences were aligned using the Codon Code Aligner programme (Version 5.0.2) and compared to sequences found on the National Centre for Biotechnology Information website (www.ncbi.nlm.nih.gov) (Abdallah et al., 2017). Ethical approvalNot required for this type of study.
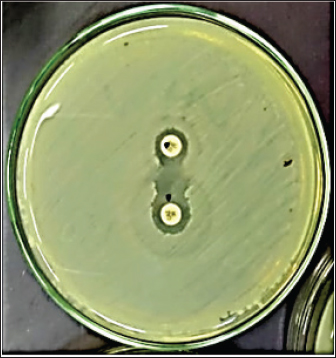
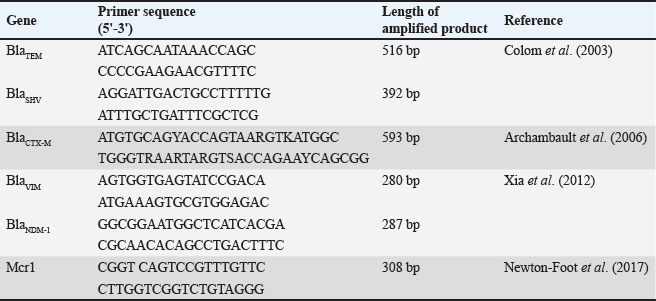
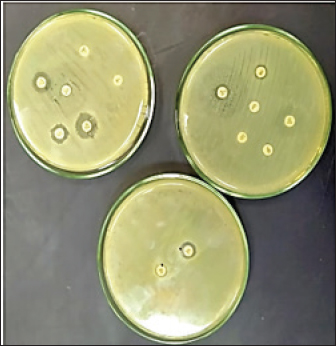
Fig. 1. Double disc synergy positive result. Table 1. Objective genes with oligonucleotide primer sequences and amplicon sizes.
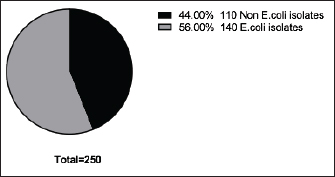
Fig. 2. Incidence of E. coli isolated from chicken farms in El-Sharkia Governorate, Egypt.
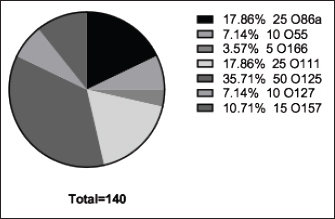
Fig. 3. The incidence of E. coli serotypes isolated from poultry samples.
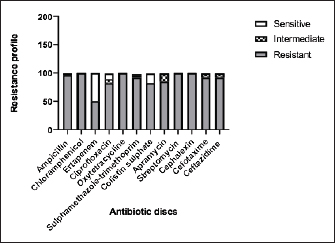
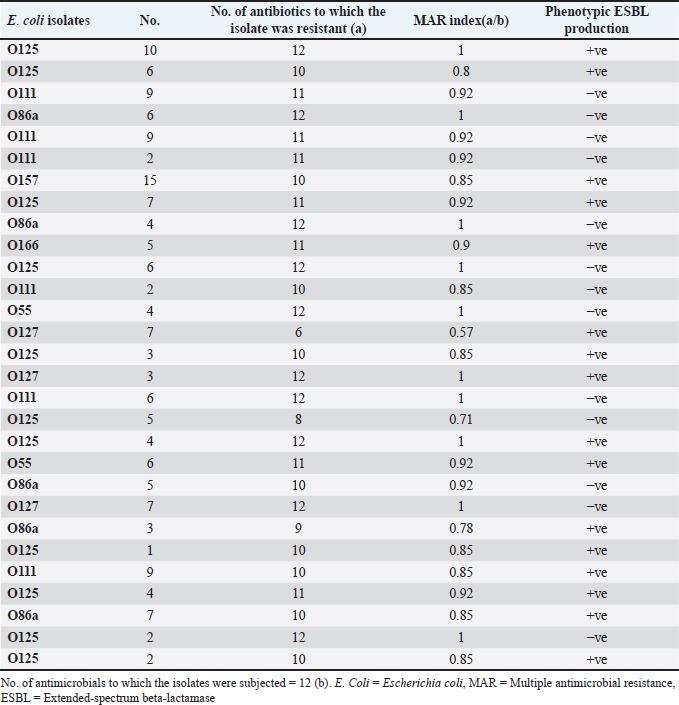
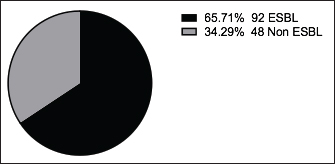
Fig. 4. Zones of inhibition of different antibiotic discs against E. coli isolate. ResultsAmong all the 250 diseased chickens, from 50 broiler poultry farms, examined for E. coli infections, 140 samples (56%) tested positive for E. coli, while 110 samples (44%) tested negative (Fig. 2). Different serovars of E. coli isolates were identified by the use of 43 monovalent group O somatic antisera (Fig. 3). The disc diffusion method was employed to evaluate the antibiotic resistance of 140 E. coli strains. The study tested the susceptibility of E. coli strains to 12 different antibiotics (Fig. 4). As depicted in Figure 5, E. coli isolates demonstrated complete resistance (100%) to cephalexin, chloramphenicol, oxytetracycline, and streptomycin, with ampicillin showing a little lower resistance rate (96%). Cefotaxime, ceftazidime, and sulfamethoxazole-trimethoprim exhibited a resistance rate of 92.9%. In addition, 115 out of the total of 140 E. coli isolates demonstrated resistance to ciprofloxacin, colistin sulfate, and apramycin. In addition, 50% of isolates showed resistance to ertapenem. All E. coli serotypes were categorized as multidrug resistant, meaning they were resistant to three or more classes of antimicrobials (Table 2). In addition, a total of 92 samples were phenotypically identified as potentially harboring ESBL (Figs. 1 and 6; Table 2).
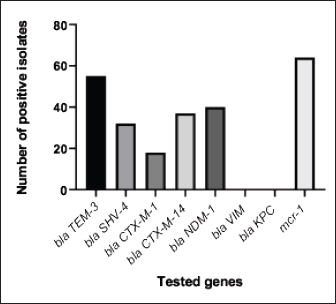
Fig. 5. The percentage of sensitivity and resistance of 140 E. coli isolates against 12 antimicrobial discs. Out of the 92 ESBL harboring E. coli, 59.7% (55 strains) were discovered to contain the blaCTX-M gene, with blaCTX-M-1 accounting for 19.6% and blaCTX-M-14 accounting for 40.2%. In the same manner, the blaTEM-3 gene was detected in 60% of the isolates, whereas 34.7% (32 out of 92) harbored the blaSHV-4 gene. With significant implications for public health, 43.4% and 69.5% of ESBL isolates were found to have the blaNDM-1 and mcr-1 genes, respectively. These data are depicted in Figure 7. DiscussionThis research study examined the prevalence of multidrug-resistant E. coli isolated from the respiratory tract of chickens exhibiting chronic respiratory illness symptoms in poultry farms located in the eastern regions of the Egyptian delta. The study examined the resistance profiles of phenotypes and genotypes against β-lactam antibiotics, carbapenems, and colistin. These profiles pose a potential risk to public health if they are transmitted to humans. Among the 250 samples, 140 were found to be positive for the presence of E. coli with a prevalence of 56%. The rate mentioned exceeds the average incidence rate (ranging from 9.52% to 36.73%) reported by Kabir (2010), who identified the age group of 4 to 6 weeks as the most susceptible. In the United States, APEC (avian pathogenic E. coli) has been consistently found in at least 30% of chicken flocks (Johnson et al., 2008). Elevated incidence rates, such as the one observed in this study, are prevalent in developing nations such as Egypt. The study conducted by Awad et al. (2016) indicated a prevalence rate of 51.85%. In addition, the researchers obtained E. coli strains from the internal organs of 92 sick broiler chickens and from 32 cloacal swabs or fecal samples from healthy broiler chickens in Algeria (Mohamed et al., 2018). A study conducted in Pakistan found a remarkably high occurrence rate of APEC isolates (89.2%) (Azam et al., 2019). The heightened occurrence of colibacillosis may be attributed to the intensive husbandry of birds in poorly ventilated housing or the presence of Mycoplasma gallisepticum infection in the flock, which leads to or exacerbates colisepticaemia. Moreover, poultry may become more susceptible to this disease due to viral infections and ecological pressures (Matuschek et al., 2018). In this study, E. coli isolates were categorized into seven distinct serotypes, with serotype O125 being the most common, accounting for 35.7% of the isolates (50 out of 140). Subsequently, there was a 17.8% occurrence rate for both O111 and O86. Additional serotypes that were found were O157 (n=15), O127 and O55 (n=10), and O166 (n=5). These findings are consistent with a prior study conducted by Rahmatallah et al. (2018), which identified O125 as the most prevalent strain of E. coli. This serogroup is responsible for 61.3% of infections in poultry, as reported by Chabou et al. (2016). Colibacillosis, a disease that affects chicken production globally, results in substantial economic losses. To treat this infection, many classes of antibiotics, including sulfonamides, aminoglycosides, β-lactams, fluoroquinolones, and tetracyclines, are commonly employed. Regrettably, this has led to the emergence of multi-resistant strains of E. coli (Grami et al., 2016). In our research, antibiotic susceptibility testing conducted on different E. coli serotypes showed that nearly all the isolates were multi-resistant, as they exhibited resistance to at least three antibiotics, indicating a MAR pattern. The E. coli isolates displayed elevated resistance to 12 antibiotics. A preceding study in Egypt examining broiler chickens found a prominent phenotypic resistance rate of E. coli to streptomycin, penicillin, trimethoprim/sulfamethoxazole, and tetracycline (Chabou et al., 2016). Other investigations have also documented the presence of multi-resistant E. coli in chicken farms in Tunisia (Kabir, 2010) and Jordan (Bernasconi et al., 2017). Our findings indicate that approximately 65.7% of all E. coli isolates are phenotypically positive for ESBL. That was in the same line with the findings of Awad et al. (2020) who stated that 65.09% of E. coli isolates were shown to be producers of ESBL. Recent surveys conducted in Egypt have revealed varying rates of phenotypically positive ESBL harboring E. coli, including 46.7% (Borowiak et al., 2017) and 75% (Badr et al., 2022a). ESBL-carrying E. coli was isolated from the intestines of approximately 34% of broilers in Sweden (Börjesson et al., 2013) and from 48.8% of retail chicken meat markets in Malaysia (Aliyu et al., 2016). Table 2. MAR index analysis and phenotypic ESBL production of E. coli serotypes.
PCR analysis and sequencing verified that out of the 92 ESBL phenotypes examined, 55 (59.7%), 32 (34.7%), 18 (19.6%), and 37 (40.2%) isolates harbor blaTEM-3, blaSHV-4, blaCTX-M-1, blaCTX-M-14 ESBL genes, respectively. The CTX-M family of genes provides resistance to cefotaxime, which is a frequently utilized antibiotic in veterinary medicine. The presence of this family has been documented in farm animals and hospitalized patients in Egypt (Braun et al., 2016), as well as in numerous food chain animals in different countries (Hartmann et al., 2012). Additional research conducted in Egypt has also documented a higher occurrence of blaCTX-M genes in ESBL-carrying E. coli from poultry offal samples (El-Shazly et al., 2017). The blaCTX-M variants, such as blaCTX-M-1 (the most common variant), blaCTX-M-2, blaCTX-M-14, and blaCTX-M-15, were previously found in hens (Randall et al., 2011). These pose a potential risk to human health, as bla CTX-M genes have already been detected in human E. coli isolates in Egypt (Mohamed Al-Agamy et al., 2006). In addition, our data indicated a high occurrence of blaTEM-3 and blaSHV-4 genes in ESBL-producing E. coli which was 55 (59.7%) and 32 (34.7%), correspondingly. In a separate investigation conducted in Egypt, the blaSHV and blaTEM genes were identified in 23.58% (7 out of 30) and 57.55% (18 out of 31) of the E. coli samples, respectively (Abdallah et al., 2015). Another investigation has shown a significant prevalence of 88.2% (27 out of 31) of blaTEM in ESBL-carrying E. coli (Colom et al., 2003). On the other hand, the blaTEM gene was found at a comparatively lower rate of 9.1% (4 out of 44) (Ibrahim et al., 2019), while the blaSHV gene was revealed in just 1.8% (2 out of 111) of the isolates (Huijbers et al., 2015). Recently, a study was conducted to analyze of E. coli chicken isolates which are associated with ESBL genes from different locations in Egypt. The study found that the EL-Sharqia governorate frequently showed the presence of blaTEM and blaCTX-M-9 genes. The Dakahlia governorate exhibited the presence of blaTEM, blaSHV, blaCTX-M-1, and blaCTX-M-9 genes, either in combination or individually, among the isolates. Conversely, isolates in Giza governorate frequently possess the blaTEM gene (Badr et al., 2022a).
Fig. 6. The phenotypic ESBL producers among 140 E. coli isolates.
Fig. 7. The incidence of antibiotic-resistant genes among 92 phenotypic ESBL-producing E. coli. An increased occurrence of CRE, including K. pneumoniae, E. coli, and P. mirabilis, has been observed in broiler farms that carried blaNDM genes. The most prevalent variants of these genes in chicken farm environments are blaNDM-1 and blaNDM-5 (Liakopoulos et al., 2018). This research identified a significant occurrence of the carbapenem-resistant gene, blaNDM-1, which was found to coexist with ESBL and colistin-resistant genes in E. coli strains obtained from chickens. The elevated occurrence of CRE in agriculture is mostly attributed to the extensive utilization of antibiotics, inadequate oversight, transportation practices, and inadequate farm sanitation. Carbapenem-resistant strains, including Acinetobacter lwoffii carrying the blaNDM-1 gene, were identified in chicken farms, duck farms, and a pig abattoir in China in 2010. The presence of P. mirabilis, a bacterium capable of harboring NDM, was documented in broiler chickens in 2021 (Xie et al., 2021). A recent study conducted in China documented the prevalence of carbapenem-resistant E. coli, P. mirabilis, and K. pneumoniae in broiler fattening farms (Su et al., 2023). In 2016, 42% of K. pneumoniae isolates from Egyptian poultry farms were carbapenem-resistant and carried blaNDM, blaKPC, and, blaOXA-48 genes (Hamza et al., 2016). A study conducted in China discovered NDM variants, specifically blaNDM-1, blaNDM-5, and blaNDM-9, in a total of 279 NDM-producing bacteria. These bacteria belong to the families Enterobacteriaceae, Alcaligenes faecalis, and Morganellaceae. The study also discovered the occurrence of mcr-8 or mcr-1 alongside blaNDM-1 in K. pneumoniae (Zhai et al., 2020). A total of 155 carbapenem-resistant isolates were identified in Egyptian research on retail chicken flesh. The analysis detected the presence of blaNDM-5 and other genes in K. pneumoniae ST147, E. coli ST648, and P. mirabilis (Shi et al., 2021). ST648 E. coli with the blaNDM-1 gene has been found in the United Kingdom, Australia, and India, together with the ST147 K. pneumoniae that has acquired the same gene has been identified in Tunisia, Iraq, and Egypt (Sadek et al., 2020). In 2019, a study conducted in China reported the existence of P. mirabilis bacteria carrying the bla NDM-1 gene in broiler chickens at slaughterhouses. In 2021, a study conducted in Egypt discovered the presence of P. mirabilis in duck farms and were shown to carry three resistant genes, namely blaOXA-1, blaNDM-1, and blaKPC (Algammal et al., 2021). Regarding colistin, 82.1% of the E. coli isolates showed resistance as revealed by AST. Prior research in Egypt has documented different degrees of colistin resistance in E. coli bacteria in the Northern governorates. One study found a high prevalence rate of 92.31% in 54 chicken farms (Awad et al., 2020), while another study revealed a low prevalence rate of 7.9% among 48 chicken farms (Moawad et al., 2018). In our investigation, we also investigated the presence of mcr1 genes, which are responsible for the emergence of colistin resistance. Among the 92 ESBL-producing E. coli isolates, 64 (69.5%) were found to possess the mcr-1 gene. Similarly, Egyptian researchers employed standard PCR assays to examine the existence of mcr1–5 gene variants in 56 phenotypically colistin-resistant E. coli samples. The results revealed that the mcr-1 gene was the sole gene present in all isolates, while the other genes (mcr-2, mcr-3, mcr-4, and mcr-5) associated with colistin resistance were not identified (Badr et al., 2022b). The presence of the mcr-1 gene is regarded as the primary determinant of colistin resistance, consistent with previous studies (Barbieri et al., 2017; Moawad et al., 2018). Consistent results have been reported throughout Northern Africa, specifically in Morocco, Algeria, and Tunisia, as well as worldwide (Bernasconi et al., 2017; Chabou et al., 2016; Grami et al., 2016; Rahmatallah et al., 2018). ConclusionThis work examined the occurrence of multidrug-resistant E. coli on Egyptian poultry farms and highlighted the resistance profiles, particularly for ESBL, carbapenems, and colistin. The results showed a higher-than-average incidence rate, which is coherent with trends observed in developing countries. The study identified multiple E. coli serotypes, including those with zoonotic potential, and observed multidrug resistance against various antibiotics. Furthermore, the increased prevalence of blaCTX-M, blaTEM, blaSHV, blaNDM-1, and mcr-1 genes in E. coli isolates poses a significant threat to public health in the respective community. This is because carbapenems and colistin represent the final lines of defense against bacteria that are resistant to multiple antibiotics. Ultimately, adopting a proactive "One Health" strategy in Egypt is crucial to prevent the rapid proliferation of antibiotic-resistant bacteria. AcknowledgmentsThe authors would like to express deep gratitude to the administration of the Faculty of Veterinary Medicine, Zagazig University, for allowing them to use all laboratories, facilities, and chemicals needed to accomplish this work. Conflict of interestThe authors declare that they have no competing interests. Authors’ contributionsRIA: Conceptualized the study, executed all experiments, and participated in the writing of the manuscript. EAA and MAK: Supervised the study, validated the results, abstracted the work, and reviewed and edited the manuscript. SKM and HHM: Analyzed the data and drafted and revised the manuscript. GAS: Conceived the work, shared in performing the experiments, wrote the manuscript, and curated and analyzed the data. All authors have read, reviewed, and approved the final manuscript. FundingThe authors did not receive any funds for this study. Data availabilityAll data are provided in the manuscript. ReferencesAbdallah, H.M., Alnaiemi, N., Reuland, E.A., Wintermans, B.B., Koek, A., Abdelwahab, A.M., Samy, A., Abdelsalam, K.W. and Vandenbroucke-Grauls, C.M.J.E. 2017. Fecal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: a hospital -based cross-sectional study. Antimicrob. Resist. Infect. Control 6, 1–7. Abdallah, H.M., Reuland, E.A., Wintermans, B.B., Al Naiemi, N., Koek, A., Abdelwahab, A.M., Ammar, A.M., Mohamed, A.A. and Vandenbroucke-Grauls, C.M.J.E. 2015. Extended-spectrum β-lactamases and/or carbapenemases-producing enterobacteriaceae isolated from retail chicken meat in Zagazig, Egypt. PLoS One 10, e0136052. Algammal, A.M., Hashem, H.R., Alfifi, K.J., Hetta, H.F., Sheraba, N.S., Ramadan, H. and El-Tarabili, R.M. 2021. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 11, 1–15. Aliyu, A.B., Saleha, A.A., Jalila, A. and Zunita, Z. 2016. Risk factors and spatial distribution of extended spectrum β-lactamase-producing-Escherichia coli at retail poultry meat markets in Malaysia: a cross-sectional study. BMC Public Health 16, 1–9. Ambler, R.P. 1980. The structure of β-lactamases. Philosophical Transactions of the Royal Society of London. B, Biol. Sci. 289, 321–331. Anago, E., Ayi-Fanou, L., Akpovi, C.D., Hounkpe, W.B., Agassounon-Djikpo Tchibozo, M., Bankole, H.S. and Sanni, A. 2015. Antibiotic resistance and genotype of beta-lactamase producing Escherichia coli in nosocomial infections in Cotonou, Benin. Ann. Clin. Microbiol. Antimicrob. 14, 1–6. Archambault, M., Petrov, P., Hendriksen, R.S., Asseva, G., Bangtrakulnonth, A., Hasman, H., and Aarestrup, F.M. 2006. Molecular characterization and occurrence of extended-spectrum β-lactamase resistance Genes among Salmonella enterica Serovar corvallis from Thailand, Bulgaria, and Denmark. Microb. Drug Resist. 12, 192–198. Awad, A., Arafat, N. and Elhadidy, M. 2016. Genetic elements associated with antimicrobial resistance among avian pathogenic Escherichia coli. Ann. Clin. Microbiol. Antimicrob. 15, 59. Awad, A.M., El-shall, N.A., Khalil, D.S., El-hack, M.E.A., Swelum, A.A., Mahmoud, A.H., Ebaid, H., Komany, A., Sammour, R.H. and Sedeik, M.E. 2020. Incidence, pathotyping, and antibiotic susceptibility of avian pathogenic Escherichia coli among diseased broiler chicks. Pathogens 9, 114. Azam, M., Mohsin, M., Sajjad-ur-Rahman, S.R. and Saleemi, M.K. 2019. Virulence-associated genes and antimicrobial resistance among avian pathogenic Escherichia coli from colibacillosis affected broilers in Pakistan. Trop. Anim. Health Prod. 51, 1259–1265. Badr, H., Reda, R.M., Hagag, N.M., Kamel, E., Elnomrosy, S.M., Mansour, A.I., Shahein, M.A., Ali, S.F. and Ali, H.R. 2022a. Multidrug-resistant and genetic characterization of extended-spectrum beta-lactamase-producing E. coli recovered from chickens and humans in Egypt. Animals 12, 346. Badr, H., Samir, A., El-Tokhi, E.I., Shahein, M.A., Rady, F.M., Hakim, A.S., Fouad, E.A., El-Sady, E.F., Ali, S.F., 2022b. Phenotypic and genotypic screening of colistin resistance associated with emerging pathogenic Escherichia coli isolated from poultry. Vet. Sci. 9, 282. Barbieri, N.L., Nielsen, D.W., Wannemuehler, Y., Cavender, T., Hussein, A., Yan, S.G., Nolan, L.K. and Logue, C.M., 2017. mcr-1 identified in avian pathogenic Escherichia coli (APEC). PLoS One 12, e0172997. Bernasconi, O.J., Principe, L., Tinguely, R., Karczmarek, A., Perreten, V., Luzzaro, F. and Endimiani, A. 2017. Evaluation of a new commercial microarray platform for the simultaneous detection of β-lactamase and mcr-1 and mcr-2 genes in Enterobacteriaceae. J. Clin. Microbiol. 55, 3138–3141. Börjesson, S., Egervärn, M., Lindblad, M. and Englunda, S. 2013. Frequent occurrence of extended-spectrum beta-lactamase- and transferable AMPC beta-lactamase-producing Escherichia coli on domestic chicken meat in Sweden. Appl. Environ. Microbiol. 79, 2463–2466. Borowiak, M., Fischer, J., Hammerl, J.A., Hendriksen, R.S., Szabo, I. and Malorny, B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72, 3317–3324. Braun, S.D., Ahmed, M.F.E., El-Adawy, H., Hotzel, H., Engelmann, I., Wei, D., Monecke, S. and Ehricht, R. 2016. Surveillance of extended-spectrum beta-lactamase-producing Escherichia coli in dairy cattle farms in the nile delta, Egypt. Front. Microbiol 7, 194821. Bubpamala, J., Khuntayaporn, P., Thirapanmethee, K., Montakantikul, P., Santanirand, P. and Chomnawang, M.T. 2018. Phenotypic and genotypic characterizations of extended-spectrum beta-lactamase-producing Escherichia coli in Thailand. Infect. Drug Resist. 11, 2151–2157. Cardozo, M.V., Liakopoulos, A., Brouwer, M., Kant, A., Pizauro, L.J.L., Borzi, M.M., Mevius, D. and de Ávila, F.A. 2021. Occurrence and molecular characteristics of extended-spectrum beta-lactamase-producing enterobacterales recovered from chicken, chicken meat, and human infections in Sao Paulo State, Brazil. Front. Microbiol. 12, 628738. Chabou, S., Leangapichart, T., Okdah, L., Le Page, S., Hadjadj, L. and Rolain, J.M. 2016. Real-time quantitative PCR assay with Taqman® probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect. 13, 71–74. Colom, K., Pérez, J., Alonso, R., Fernández-Aranguiz, A., Lariño, E. and Cisterna, R. 2003. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA–1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 223, 147–151. Coppola, N., Freire, B., Umpiérrez, A., Cordeiro, N.F., Ávila, P., Trenchi, G., Castro, G., Casaux, M.L., Fraga, M., Zunino, P., Bado, I. and Vignoli, R. 2020. transferable resistance to highest priority critically important antibiotics for human health in Escherichia coli strains obtained from livestock feces in Uruguay. Front. Vet. Sci. 7, 588919. El-Shazly, D.A., Nasef, S.A., Mahmoud, F.F. and Jonas, D. 2017. Expanded spectrum β–lactamase producing Escherichia coli isolated from chickens with colibacillosis in Egypt. Poult. Sci. 96, 2375–2384. Giufrè, M., Mazzolini, E., Cerquetti, M., Brusaferro, S., Accogli, M., Agnoletti, F., Agodi, A., Alborali, G.L., Arghittu, M., Auxilia, F., Barchitta, M., Bosco, N., Camporese, A., Carfora, V., Collini, L., D’Agaro, P., De Rosa, R., Formenti, N., Franco, A., Koncan, R., Lanzafame, P., Mazzariol, A., Moschioni, C., Pane, S., Putignani, L. and Thoma, C. 2021. Extended-spectrum β-lactamase-producing Escherichia coli from extraintestinal infections in humans and from food-producing animals in Italy: a “One Health” study. Int. J. Antimicrob. Agents 58, 106433. Grami, R., Mansour, W., Mehri, W., Bouallègue, O., Boujaâfar, N., Madec, J. and Haenni, M. 2016. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, tunisia, july 2015. Eurosurveillance 21, 1–5. Hamza, E., Dorgham, S.M. and Hamza, D.A. 2016. Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. J. Glob. Antimicrob. Resist. 7, 8–10. Hartmann, A., Locatelli, A., Amoureux, L., Depret, G., Jolivet, C., Gueneau, E. and Neuwirth, C. 2012. Occurrence of CTX-M producing Escherichia coli in soils, cattle, and farm environment in France (Burgundy region). Front. Microbiol. 3, 18761. Huijbers, P.M.C., Blaak, H., De Jong, M.C.M., Graat, E.A.M., Vandenbroucke-Grauls, C.M.J.E. and De Roda Husman, A.M. 2015. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ. Sci. Technol. 49, 11993–12004. Ibrahim, R.A., Cryer, T.L., Lafi, S.Q., Basha, E.A., Good, L. and Tarazi, Y.H. 2019. Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors. BMC Vet. Res. 15, 1–16. Johnson, T.J., Wannemuehler, Y., Doetkott, C., Johnson, S.J., Rosenberger, S.C. and Nolan, L.K. 2008. Identification of minimal predictors o f avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 46, 3987–3996. Kabir, S.M.L. 2010. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int. J. Environ. Res. Public Health 7, 89–114. Liakopoulos, A., Van Der Goot, J., Bossers, A., Betts, J., Brouwer, M.S.M., Kant, A., Smith, H., Ceccarelli, D. and Mevius, D. 2018. Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 8, 1–13. Matuschek, E., Åhman, J., Webster, C. and Kahlmeter, G. 2018. Antimicrobial susceptibility testing of colistin—evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin. Microbiol. Infect. 24, 865–870. Mendelson, M., Brink, A., Gouws, J., Mbelle, N., Naidoo, V., Pople, T., Schellack, N., van Vuuren, M., Rees, H., Banoo, S., Bokaba, K., Collins, M., Faure, K., Herbst, M., Hoek, B., Lancaster, R., Lotter, J., Modisane, M., Mohlala, M., Mokantla, E., Molatuli, A., Molefe, M., Molewa, G., Mompati, K., Moshiga, L., Munbodh, S., Nkambule, P., Patterson, C., Reddy, D., Sigobondhla, A., Suliman, S. and Swan, G. 2018. The one health stewardship of colistin as an antibiotic of last resort for human health in South Africa. Lancet Infect. Dis. 18, e288–e294. Moawad, A.A., Hotzel, H., Awad, O., Tomaso, H., Neubauer, H., Hafez, H.M. and El-Adawy, H., 2017. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 9, 1–13. Moawad, A.A., Hotzel, H., Neubauer, H., Ehricht, R., Monecke, S., Tomaso, H., Hafez, H.M., Roesler, U. and El-Adawy, H. 2018. Antimicrobial resistance in Enterobacteriaceae from healthy broilers in Egypt: emergence of colistin-resistant and extended-spectrum β-lactamase-producing Escherichia coli. Gut Pathog. 10, 1–12. Mohamed Al-Agamy, M.H., El-Din Ashour, M.S. and Wiegand, I. 2006. First description of CTX-M β-lactamase-producing clinical Escherichia coli isolates from Egypt. Int. J. Antimicrob. Agents 27, 545–548. Mohamed, L., Ge, Z., Yuehua, L., Yubin, G., Rachid, K., Mustapha, O., Junwei, W. and Karine, O. 2018. Virulence traits of avian pathogenic (APEC) and fecal (AFEC) E. coli isolated from broiler chickens in Algeria. Trop. Anim. Health Prod. 50, 547–553. Newton-Foot, M., Snyman, Y., Maloba, M.R.B. and Whitelaw, A.C. 2017. Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. clinical isolates from the Western Cape region of South Africa. Antimicrob. Resist. Infect. Control 6, 1–7. Osei Sekyere, J., 2019. Mcr colistin resistance gene: a systematic review of current diagnostics and detection methods. Microbiologyopen 8, e00682. Paul, M., Daikos, G.L., Durante-Mangoni, E., Yahav, D., Carmeli, Y., Benattar, Y.D., Skiada, A., Andini, R., Eliakim-Raz, N., Nutman, A., Zusman, O., Antoniadou, A., Pafundi, P.C., Adler, A., Dickstein, Y., Pavleas, I., Zampino, R., Daitch, V., Bitterman, R., Zayyad, H., Koppel, F., Levi, I., Babich, T., Friberg, L.E., Mouton, J.W., Theuretzbacher, U. and Leibovici, L. 2018. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: an open-label, randomised controlled trial. Lancet Infect. Dis. 18, 391–400. Poirel, L., Madec, J.-Y., Lupo, A., Schink, A.-K., Kieffer, N., Nordmann, P. and Schwarz, S. 2018. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6(4), doi:10.1128/MICROBIOLSPEC.ARBA-0026-2017. Rahmatallah, N., El Rhaffouli, H., Laraqui, A., Sekhsokh, Y., Lahlou-Amine, I., El Houadfi, M. and Fihri, O.F. 2018. Detection of colistin encoding resistance genes MCR-1 in isolates recovered from broiler chickens in morocco. Saudi J. Pathol. Microbiol. 3(12), 520–521. Randall, L.P., Clouting, C., Horton, R.A., Coldham, N.G., Wu, G., Clifton-Hadley, F.A., Davies, R.H. and Teale, C.J. 2011. Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J. Antimicrob. Chemother. 66, 86–95. Sadek, M., Poirel, L., Nordmann, P., Nariya, H., Shimamoto, T. and Shimamoto, T. 2020. Genetic characterisation of NDM-1 and NDM-5-producing Enterobacterales from retail chicken meat in Egypt. J. Glob. Antimicrob. Resist. 23, 70–71. Shi, X., Li, Y., Yang, Y., Shen, Z., Cai, C., Wang, Y., Walsh, T.R., Shen, J., Wu, Y. and Wang, S. 2021. High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J. Hazard Mater. 406, 124298. Su, Y., Xin, L., Zhang, F., Peng, C., Li, Z., Liu, C. and Wang, F. 2023. Drug resistance analysis of three types of avian-origin carbapenem-resistant Enterobacteriaceae in Shandong Province, China. Poult. Sci. 102, 102483. Swayne, D.E. 1998. Laboratory manual for the isolation and identification of avian pathogens. American Association of Avian Pathologists, University of Pennsylvania. Taggar, G., Rheman, M.A., Boerlin, P. and Diarra, M.S. 2020. Molecular epidemiology of carbapenemases in enterobacteriales from humans, animals, food and the environment. Antibiotics 9, 693. White, A. and Hughes, J.M. 2019. Critical importance of a one health approach to antimicrobial resistance. Ecohealth 16, 404–409. Xia, Y., Liang, Z., Su, X. and Xiong, Y. 2012. Characterization of carbapenemase genes in enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a University Hospital in Chongqing, China. Ann. Lab. Med. 32, 270–275. Xie, X., Zhang, J., Wang, H.N. and Lei, C.W. 2021. Whole genome sequence of a New Delhi metallo-β-lactamase 1-producing Proteus mirabilis isolate SNYG35 from broiler chicken in China. J. Glob. Antimicrob. Resist. 24, 266–269. Zhai, R., Fu, B., Shi, X., Sun, C., Liu, Z., Wang, S., Shen, Z., Walsh, T.R., Cai, C., Wang, Y. and Wu, C., 2020. Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ. Int. 139, 105715. | ||
| How to Cite this Article |
| Pubmed Style Ali RI, Abdelaziz EA, Kamel MA, Murad SK, Abdallah HM, Salem GA. Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 459-469. doi:10.5455/OVJ.2024.v14.i1.42 Web Style Ali RI, Abdelaziz EA, Kamel MA, Murad SK, Abdallah HM, Salem GA. Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt. https://www.openveterinaryjournal.com/?mno=181966 [Access: July 01, 2025]. doi:10.5455/OVJ.2024.v14.i1.42 AMA (American Medical Association) Style Ali RI, Abdelaziz EA, Kamel MA, Murad SK, Abdallah HM, Salem GA. Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 459-469. doi:10.5455/OVJ.2024.v14.i1.42 Vancouver/ICMJE Style Ali RI, Abdelaziz EA, Kamel MA, Murad SK, Abdallah HM, Salem GA. Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt. Open Vet J. (2024), [cited July 01, 2025]; 14((1) (Zagazig Veterinary Conference)): 459-469. doi:10.5455/OVJ.2024.v14.i1.42 Harvard Style Ali, R. I., Abdelaziz, . E. A., Kamel, . M. A., Murad, . S. K., Abdallah, . H. M. & Salem, . G. A. (2024) Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 459-469. doi:10.5455/OVJ.2024.v14.i1.42 Turabian Style Ali, Reem I., El-sayed A. Abdelaziz, Mohammed A. Kamel, Susan K. Murad, H. M. Abdallah, and Gamal A. Salem. 2024. Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 459-469. doi:10.5455/OVJ.2024.v14.i1.42 Chicago Style Ali, Reem I., El-sayed A. Abdelaziz, Mohammed A. Kamel, Susan K. Murad, H. M. Abdallah, and Gamal A. Salem. "Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt." Open Veterinary Journal 14 (2024), 459-469. doi:10.5455/OVJ.2024.v14.i1.42 MLA (The Modern Language Association) Style Ali, Reem I., El-sayed A. Abdelaziz, Mohammed A. Kamel, Susan K. Murad, H. M. Abdallah, and Gamal A. Salem. "Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 459-469. Print. doi:10.5455/OVJ.2024.v14.i1.42 APA (American Psychological Association) Style Ali, R. I., Abdelaziz, . E. A., Kamel, . M. A., Murad, . S. K., Abdallah, . H. M. & Salem, . G. A. (2024) Phenotypic and genotypic characterization of extended spectrum beta-lactamase producing E. coli harboring carbapenem and colistin resistant genes from poultry farms in Egypt. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 459-469. doi:10.5455/OVJ.2024.v14.i1.42 |