| Research Article | ||
Open Vet J. 2023; 13(7): 879-893 Open Veterinary Journal, (2023), Vol. 13(7): 879-893 Original Research Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing processHannes Beims1,2,3*, Martina Janke2, Werner von der Ohe2 and Michael Steinert31Bezirk Oberbayern, Fachberatung für Imkerei, München, Germany 2Lower Saxony State Office for Consumer Protection and Food Safety, Institute for Apiculture, Celle, Germany 3Institut für Mikrobiologie, Technische Universität Braunschweig, Braunschweig, Germany *Corresponding Author: Hannes Beims. Bezirk Oberbayern, Fachberatung für Imkerei, München, Germany. Email: Hannes.Beims [at] Bezirk-Oberbayern.de Submitted: 31/03/2023 Accepted: 21/06/2023 Published: 31/07/2023 © 2023 Open Veterinary Journal
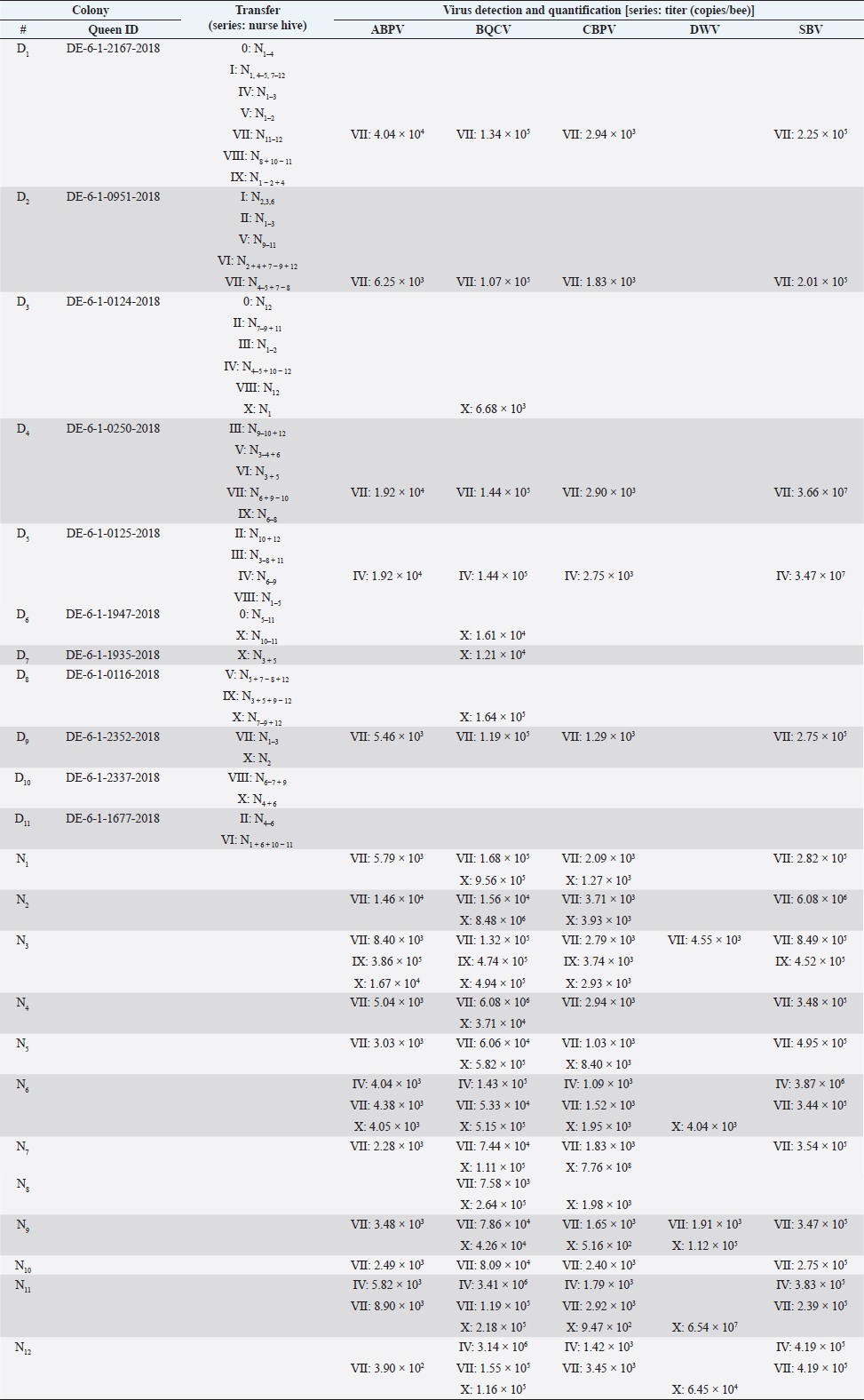
AbstractBackground: Honeybees are one of the three most important animals for mankind. In order to be safe and increase number of bee colonies for pollination, the breeding of queens is necessary. For several decades, bees were selected on economic and behavioral aspects. With the appearance of the neozootic mite Varroa destructor beekeepers were forced to adapt their methods. Varroa destructor can act as a vector for many different bee pathogenic viruses and by this potentiates its devastating impact. Aim: Methods of rearing queens were not evaluated since the mites’ appearance. Besides scientific approaches, viruses received too little attention in regard to the rearing process of honeybee queens. Herein, we present a detailed analysis of virus abundances [Aparavirus, acute bee paralysis virus (ABPV); Triatovirus, black queen cell virus (BQCV); Cripavirus, chronic bee paralysis virus (CBPV); and Iflaviruses, deformed wings virus (DWV), Sacbrood virus (SBV), VDV-1] in breeding hives, donating first instar larvae, hives that are nursing these larvae until the pupa stage, and on queens of Apis mellifera in a breeding apiary. Methods: Nurse and donor colonies of the queen-rearing process were sampled in the year 2020 and analyzed by RT qPCR. Virus quantifications were correlated with queen mortalities and seasonal effects. Results: Virus detections increased in reared queens, however, the elevated virus titers did not increase the mortality of the queens until their exclosure. Moreover, we observed a lower interrelation between virus abundance in queens and their original donor colonies, than between nurse hives and their nursed queens. Conclusion: The bee pathogenic viruses ABPV, BQCV, CBPV, DWV, SBV, and VDV-1 do not influence the mortality of bee queens during the rearing process. Whether respective virus loads result in sublethal or long-term effects remains to be elucidated. Keywords: Apis mellifera, Breeding, Virus abundance, Optimizing breeding procedures. IntroductionThe western honeybee Apis mellifera is one of the most important animals for mankind, because of its role in pollination. Moreover, the production of honey and other bee products represents an economic aspect of beekeeping (Ritten et al., 2018). However, since the appearance of the neozootic mite Varroa destructor (Traynor et al., 2020), beekeepers all over the world have had to adapt their methods to reduce the risk of Varroosis (van Alphen and Fernhout, 2020). Different pathogenic bee viruses are known (McMenamin and Genersch, 2015; Tantillo et al., 2015) and V. destructor can act as a vector for the majority of them (Posda-Florez et al., 2020). The most devastating bee viruses are deformed wing virus (DWV) (McMenamin and Flenniken, 2018) and chronic bee paralysis virus (CBPV) (Amiri et al., 2014) of which the latter is not transmitted by V. destructor (De Miranda et al., 2013). DWV can be subdivided into several groups (DWV-A, DWV-B, DWV-C) (Kevill et al., 2017). DWV-B, also known as VDV-1, mainly causes deformed wings in A. mellifera and decreased life expectancy, leading to winter losses in combination with V. destructor infestation (Gisder et al ., 2009; Gisder and Genersch, 2020). CBPV is less understood, symptoms can be detected in single bees or affect complete hives. Infected bees tremble, often lose their hair, which makes them look black, and are unable to coordinate their wings (Ribière et al., 2010). CBPV can easily be transmitted by contact through micro wounds (i.e. broken chitin hairs) between bees (Amiri et al., 2014). Importantly, most viruses [i.e. the AKI complex of acute bee paralysis virus (ABPV), Israeali acute paralysis virus, and Kashmir bee virus (KBV), as well as other viruses, like black queen cell virus (BQCV) and Sacbrood virus (SBV)] can be transmitted vertically and horizontally (Amiri et al., 2014; Ryabov et al., 2016). However, these viruses can cause several, unspecific symptoms, but their impact on colonies’ health is less devastating than DWV and CBPV (De Miranda et al., 2013; McMenamin and Genersch, 2015; Ravoet et al., 2015; Schurr et al., 2019). In order to increase the economic efficiency and health of honey bees, honeybee colonies are valued according to different morphometric criteria (Deutscher Imkerbund, 2017), i.e., cubital index (Beims et al., 2020a), and selected on behavioral criteria, i.e., colony strength and efficiency in collecting nectar, and for breeding (Ravoet et al., 2015; Tiesler et al., 2016). Some apiaries are specialized in rearing bee queens, which are mated at mating stations with selected drones (Tiesler et al., 2016). However, the majority of breeding methods harbor the risk of virus transfer between different colonies, castes, and individuals. The LAVES institute for apiculture rears more than 2,000 queens per season by using nurse hives. These nurse hives are used during the entire breeding season and kept vital by adding capped brood frames from unspecific colonies. Hatched virgin queens are transferred into nukes and mated on mating stations. This procedure harbors the risk of virus transmission at several points in the process of queen production. In the present study we (i) monitored donor and nurse colonies for virus infections, (ii) identified critical steps for virus transmission during the breeding processes and (iii) paved the way for alternate breeding methods which minimize the risk of virus transmission. Materials and MethodsRearing of queen beesPre-selected colonies of A. mellifera subsp. carnica, belonging to the Troisek line (C-T) of the Institute for Apiculture Celle were used as donor colonies (nD=11). Queens of these colonies are defined as 2a ancestry, according to Goetze (1936). First instar larvae at the age of less than 24 hours were grafted into artificial queen cells and transferred into nurse colonies without a queen (nN=12) (Table 1). These larvae represent queens, called 1a (Goetze, 1936). Weekly, each nurse colony received 42 cells per series. In total, 11 series (0–X) were performed in 2020 (Table 1). Nurse colonies were inspected for unselected queen cells and added with capped brood frames from un-screened, random colonies, weekly. Queen cells were transferred after capping into an incubator (IN110, Merck, Germany) and cultivated until they hatched at 35°C. Two to six different donor colonies were used per series. Each nurse colony received only larvae from one donor colony per series (Table 1). The difference between transferred larvae and accepted cells was defined as mortality before capping and the difference between hatched queens and accepted cells as mortality after capping. Sampling for virus detection and disease screeningColonies were optically inspected for symptoms of different diseases (i.e. Varroosis, Chalkbrood, American Foulbrood) and sampled by catching and screening of 15 workers before the breeding season 2020. Weekly, 15 workers were sampled from donor and nurse colonies. Queens which have not hatched in time or died in their cells were analyzed for viruses individually. Preparation of virus RNAHeads of sampled individuals were pooled per colony and transferred into lysis tubes (MN Bead Tube Type G, Macherey-Nagel, Germany) and lyzed in 1 ml 154 mM NaCl (Roth, Germany) for 1 minute in a tissue lyzer (SpeedMill PLUS, Analytik Jena, Germany). Homogenates were centrifuged for 2 minute at 12,000 × g, the supernatant was used for RNA extraction. Samples were extracted automatically (epMotion 5075, Eppendorf, Germany), using the NukleoMag® VET chemistry (Macherey-Nagel, Germany). RNA was finally eluted in 50 μl elution buffer (5 mM Tris/HCl, pH 8.5, supplied by the manufacturers). Detection of bee virusesViruses (ABPV, BQCV, CBPV, DWV, SBV, VDV-1) were detected by RT multiplexed qPCR (Schurr et al., 2019) using the Luna® Universal probe one-step RT-qPCR Kit (New England Biolabs, USA) in 5 μl template on an AriaMX thermocycler (Agilent, USA). Results with a quantification cycle (Cq) (ΔRn) <30 were interpreted as positive (Ruiz-Villalba et al., 2017; Beims et al., 2020b) and used for further analysis. A reference gene to normalize the qPCRs was not used (Engel et al., 2015) because of multiplexing the different dyes and an internal reference dye per reaction (Schurr et al., 2019). Statistical analysisStatistical analyses were performed by using R (version ×64 3.6.1) (R Core Team, Austria). Significance was checked by unpaired student t-test. When data were tested individually against the serial results, one sample t-test was used. Ethical approvalNot needed for this study. ResultsVirus detection in donor colonies (2a)A total number of 11 donor colonies (D1–D11) were used in 2020. Various numbers of donor colonies were used per series (Table 1). Five different virus species: ABPV, BQCV, CBPV, SBV, and DWV were screened, while species DWV was subdivided into two groups, DWV (mainly representing type DWV-A) and VDV-1 (mainly representing DWV-B) (Ravoet et al., 2015; Gisder and Genersch, 2020). Viruses were detected in three series (Fig. 1): IV (D5: ABPV, BQCV, CBPV, and SBV), VII (D1, 2, 4, 9: ABPV, BQCV, CBPV, SBV), and X (D3, 6, 7, 8, 10: BQCV). In series IV, four nurse hives (N6, N7, N8, and N9), representing 33.33% of derived larvae, were affected by the positively tested donor hive. All larvae of series VII were affected by the positively tested donor hives. Except for one nurse hive (N2), all other nurse hives in series X, representing 91.67% of derived larvae, were affected by the infected donor hives. However, only one species (SBV) of the genus Iflavirus was detected in the donor colonies. Significant differences between the viruses’ abundances were not detected. Table 1. Virus detection and quantification in donor colonies and nurse hives.
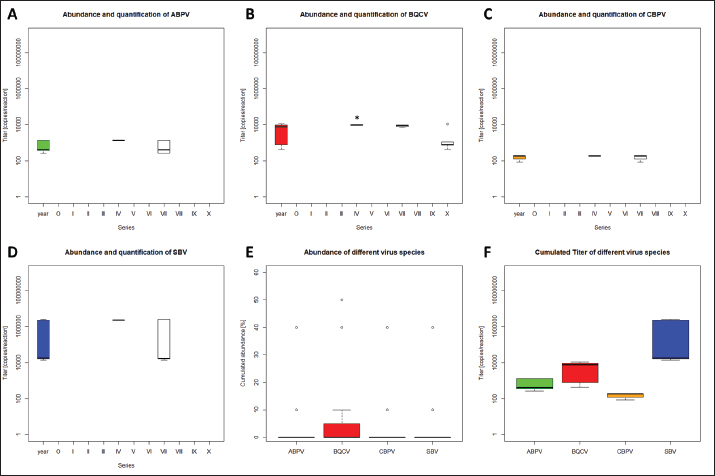
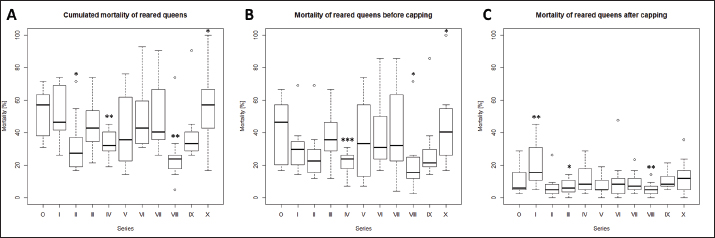
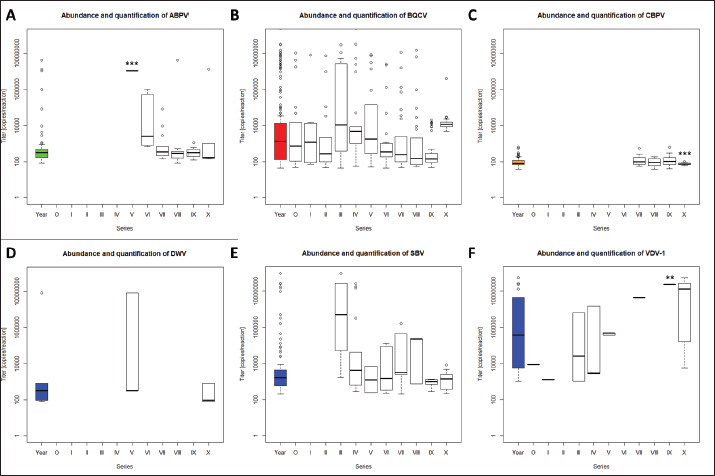
Quantification of viruses in donor colonies (2a)Titers of detected viruses were quantified (Fig. 1F). The titer of ABPV showed a median of 6,241.5 copies/bee (Min=4,035; Max=19,155; n=5). An increased median was detected for BQCV with 113,028 copies/bee (Min=6,673.5; Max=163,272 n=10) with a significant deviation of series IV (p=0.0207, n=1) compared to its cumulated titers. CBPV revealed a median of 2,750.85 copies/bee (Min=1,288.35: Max=2,940.6; n=5) and SBV with 273,930 copies/bee (Min=201,660; Max=36,608,715; n=5). Virus detection in nurse hivesTwelve nurse hives (N1–N12) were used and sampled for virus abundance, weekly (Table 1). ABPV was detected in series IV (N6, 11), VII (N1-7, 9-12), IX (N3), and X (N3, 6), resulting in 12.30% ± 8.22% affected larvae (Fig. 2A). BQCV could also be found in series IV (N6, 11, 12), VII (N1-12), IX (N3) and X (N1-9, 11-12) (Fig. 2B). In total 18.12% ± 10.61% of nursed larvae were determined BQCV-positive. Additionally, CBPV-positive nurse hives were identified in series IV (N6, 11, 12), VII (N1-7, 9-12), IX (N3), and X (N1-3, 5-9, 11) (Fig. 2C) with an effect on 18.36% ± 10.05% queen larvae. The genus Iflavirus was presented by the species DWV (VII: N3, 9; X: N6, 9, 11, 12) and SBV (IV: N6, 11, 12; VII: N1-7, 9-12; IX: N3) in the nurse hives (Fig. 2D and E). However, 4.55% ± 3.25% of queen larvae were affected by DWV and 11.54% ± 8.37% by SBV. Virus abundance and quantification in nurse hivesTiter for each virus species were quantified in the nurse hives (Fig. 2). A median of 5,418 copies/bee (Min=2,287.5; Max=386,130; n=16) was determined for ABPV. BQCV’ titer showed a median of 154,050 copies/bee (Min=7,575; Max=9,842,115; n=27), while a significant difference was calculated between the titer detected in series IX (p < 0.0001; n=1), compared to its cumulated titers. For CBPV a median of 2,025 copies/bee was determined (Min=510; Max=775,985,115; n=24). Two species were quantified for the genus Iflavirus. DWV was quantified with a titer median of 34,500 copies/bee (Min=1,905; Max=65,357,055; n=6) and SBV with 383,130 copies/bee (Min=238,620; Max=3,874,905; n=15). Significant differences in abundance and their titers between the different virus species were not detected. Mortality of queens (1a)Mortality of queens was recorded during the rearing process in the used nurse colonies (nN=12). Between the larvae transfer from donor colonies and capping in the nurse colonies, a mean of mortality was recorded (32.58% ± 1.75%) overall 11 series. Significant deviations occurred in series IV (22.42% ±2.32%, p < 0.0001), VIII (20.24% ± 4.86%, p=0.0215), and X (43.65% ± 6.41% , p=0.0374). After capping, the mortality resulted in 10.20% ± 0.81%, within significant deviations in series I (20.83% ± 3.74%, p=0.0097), III (6.55% ± 1.35%, p=0.0198) and VIII (5.36% ± 1.18%, p=0.0012). Taken together, a cumulated mortality of 42.70% ± 1.70% was recorded overall 11 series. A significant decrease in mortalities was detected in series II (32.14% ± 4.79%, p=0.0474), IV (32.34% ± 1.98%, p=0.0028) and VIII (25.60% ± 4.86%, p=0.0011). Increased mortality was detected in series X (55.16% ± 6.41%, p=0.0153) (Fig. 3).
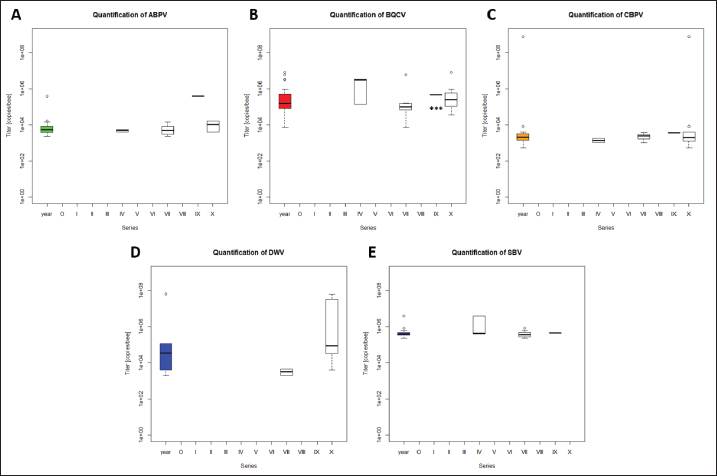
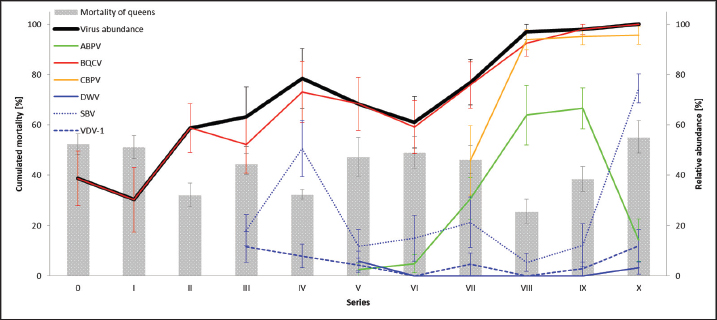
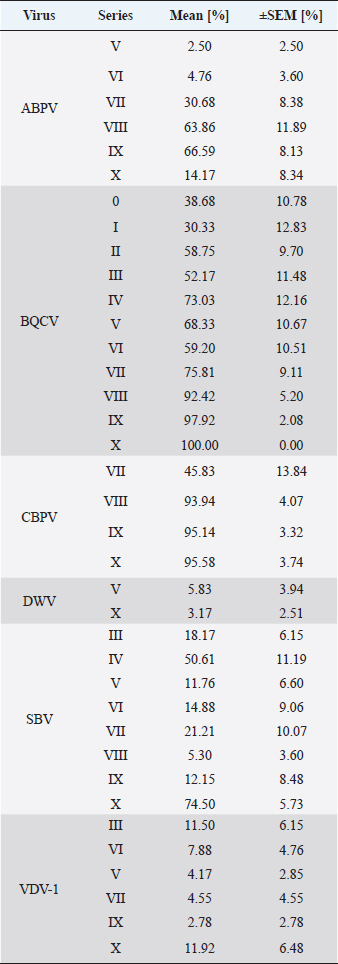
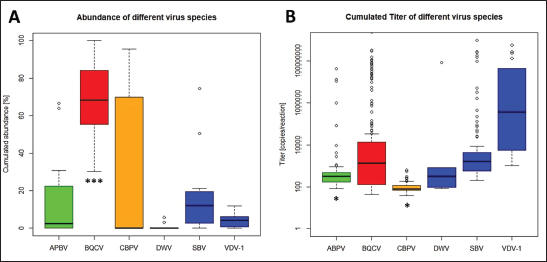
Fig. 1. Abundance and quantification of different virus species in donor colonies. Titers of detected virus species were determined (copies/bee). Colored boxplots indicate the cumulated results of series 0–X: green, Aparavirus (ABPV); red, Triatovirus (BQCV); orange, Cripavirus (CBPV); blue, Iflavirus (SBV). A–D, Abundance of virus species among series 0–X; E, relative abundance of virus species in 2020; F, cumulated titer of detected virus species in 2020. Significance code: *, 0.05> p > 0.01. Virus detection in queens (1a)Queens that died after cell capping were used for virus analysis (n=328). During the breeding season 2020, viruses were detected in 70.41% ± 3.39% of the killed queens. The initial detection quote was 38.68% ± 10.78% (series 0) and decreased to 30.33% ± 12.83% (series I), followed up by a local side lope in series IV with 78.48% ± 11.90% (II: 58.75% ± 9.70%; III: 63.17% ± 11.86%). The detection rate decreased in series V (68.33% ± 10.67%) and VI (60.98% ± 10.26%). From series VII (76.95% ± 9.01%) a continuous increase (VIII: 96.97% ± 3.03%; IX: 97.92% ± 2.08%) up to 100% in series X was detected (Fig. 4). Abundance of different virus genera in queens (1a)ABPV was detected from series V up to series X. BQCV was detected in all series. CBPV could be detected in series VII to X. All three species of Iflavirus were found in series V and X. SBV was detected in additional six series (III, IV, VI, VII, VIII, and IX). VDV-1 could be found in four of these series (III, IV, VII, and IX) and individually in two more series (0, I) (Table 2). Virus quantification in queens (1a)Additionally, to the virus abundance, its viral titer was determined (Fig. 5). ABPV was detected in six series (V–X) with a median of 321 copies/bee (Min=81; Max=42,395,275; n=86). The significant deviation was detected in series V (p < 0.0001; nV=1), according to the cumulated quantification rate. BQCV was detected in all series with a median of 13,774,845 copies/bee (Min=44; Max=244,840,507; n=315), without any significant differences between the individual series and the cumulated quantification rate. CBPV could be detected in four series (VII–X). Its quantification median was at 79.49 copies/bee (Min=38.27; Max=633.83; n=163). A significant difference according to the cumulated quantification rate was identified in series X (p < 0.0001; nX=61). The genus Iflavirus was detected in 10 of 11 series (0, I, III–X). DWV could be detected in two series (V, X) with a median of 322 copies/bee (Min=80; Max=84,990,896; n=5). SBV was detected in series III–X. Its detection rate was determined with a median of 1,612 copies/bee (Min=201; Max=984,611,011; n=96). Significant differences between the cumulated detection rates and the individual series were not detected. The third species, VDV-1, was detected in eight series (0, I, III–V, VII, IX, X). The cumulated median of copies/bee was determined on 378,820 (Min=1,050; Max=570,926,559; n=17). A significant deviation was detected in series IX (p=0.004; nIX=1). Taken together, the quantified titers of ABPV and CBPV differed significantly from other respective virus titers ( pABPV=0.0307; pCBPV=0.0179) (Fig. 6B); whereas, the percentage abundance of BQCV was significantly increased, compared to the other virus abundances ( p < 0.0001) (Fig. 6A).
Fig. 2. Abundance and quantification of different virus species in nurse hives. Titers of detected virus species were determined (copies/bee). Colored boxplots indicate the cumulated results of series 0–X: green, Aparavirus (ABPV); red, Triatovirus (BQCV); orange, Cripavirus (CBPV); blue, Iflavirus (DWV and SBV). Abundance of virus species among series 0–X: A, ABPV; B, BQCV; C, CBPV; D, DWV; E, SBV. Significance code: ***, p < 0.001.
Fig. 3. Mortality of reared queens during the breading season 2020. Mortalities [%] were calculated for each series (0–X) per used nurse hives (n=12). A, cumulated mortalities per series; B, Mortality of queen larvae in nurse hive before capping (larvae stage); C, mortality of queens after cell capping (late larvae stage, pupae and imago). Significance codes: *, 0.05> p > 0.01; **, 0.01> p > 0.001; ***, p < 0.001.
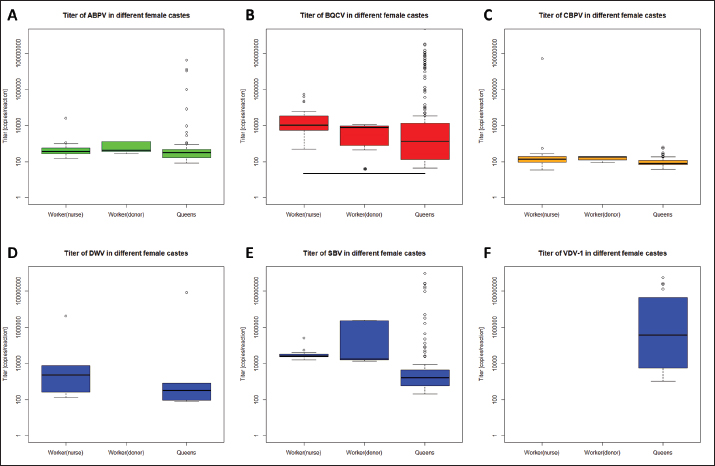
Fig. 4. Mortality of queens and virus abundances during the breading season 2020. Cumulated mortalities of reared queens were calculated (mean ± SEM) for each series (0–X, n=504 queens/series) and virus abundances [%] in dead queens (after cell capping) were determined (mean ± SEM). Virus families were colored individually: black, quote of virus-positive tested individuals; green, Aparavirus (ABPV); red, Triatovirus (BQCV); orange, Cripavirus (CBPV); blue, Iflavirus (DWV, continuous line; SBV, dotted line; VDV-1, dashed line). Barplots, cumulated mortality of queens. Correlation of virus detection and mortalityAccording to the results of virus detection, data were separated into three groups: abundance of viruses in (i) donor colonies (ii) nurse colonies, and (iii) donor and nurse colonies. Control data were presented by the absence of virus detection. The mortality of queens did not change significantly (unpaired student t-test) when viruses were detected in the donor, nurse, or both colonies. Virus titers in different female bee castesTiters of different virus species were determined separately for workers of donor colonies (2a), workers of nurse hives, and killed queens (1a). ABPV was detected in all three groups, without significant differences in the recorded titers (Fig. 7A). BQCV was found in workers and queens, with significant differences in the titer between the worker groups ( p=0.0328) (Fig. 7B). CBPV was abundant in all three tested groups of female bees with significant differences in titer between queens and worker from donor hives ( p=0.0023) (Fig. 7C). DWV was detected in workers of nurse hives and queens, but not in workers of the donor colonies. The differences in DWV titers were not significant (Fig. 7D). The titer of SBV did not differ significantly between all three tested groups (Fig. 7E). VDV-1 was only detected in queens. DiscussionSince the appearance of V. destructor in the natural distribution area of A. mellifera, beekeeping needs to be adapted to changing challenges (Traynor et al., 2020). In the first years, the parasite itself was the focus of interest (Zheguang et al., 2018), followed by a wide range of practical control measures (Rosenkranz et al., 2010; Dietemann et al., 2012; Frey and Rosenkranz, 2014) and therapeutically treatments (Dietemann et al., 2012; Ziegelmann et al., 2018). In the last two decades, however, the attention shifted towards Varroa-transmitted viruses (Gisder and Genersch, 2015), especially on DWV (Gisder et al., 2009). Since DWV was subdivided into different types (Kevill et al., 2017), DWV-B, also known as VDV-1 (Moore et al., 2011), came into the focus of A. mellifera winter losses, caused by Varroa transmitted DWV-B (Gisder and Genersch, 2020). Similarly, to DWV variants, other viruses massively affect the health of honey bees (Tantillo et al., 2015). Although, genome structure, replication and infection routes of several viruses are known (De Miranda et al., 2010; Ribière et al., 2010; Tantillo et al., 2015; Ryobov et al., 2016; Kevill et al., 2017; Spurny et al., 2017), studies addressing the infection of queens are rare or missing (Amiri et al., 2014; De Graaf et al., 2020). Except for one study on the susceptibility of queens to CBPV and another on the insemination of queens with infected sperm (Prodelalova et al., 2019) little is known about virus transmissions during breeding. Interestingly, a recent study on suppressed in ovo virus infection (SOV) indicates a new route of preventing virus transmission in breeding honeybees (De Graaf et al., 2020). In the present study, we analyzed the rearing process of honeybee queens in a well-established system, monitored by a quality management system. Table 2. Abundance of different viruses in queens.
Vertical transmission routes of virusesDuring the rearing process of queens, the donor colonies represent the step of potential vertical virus transmissions (De Graaf et al., 2020). Virus-positive queens may produce eggs that become infected by the maternal route. The paternal route of vertical transmission is given by the semination of healthy eggs with virus-positive drone sperm. Workers of donor colonies were screened before larvae donation. Each sample contained 15 randomly collected individuals. ABPV, BQCV, CBPV, and SBV could be detected, but these viruses could not be detected in the following series. Initially, we assumed to detect potential vertical transmitted virus infections in the workers of donor colonies. In the case of the maternal route, all workers should have been positive for viruses. According to the results, we can exclude the maternal route of vertical virus transmission. Focusing on the paternal way of vertical virus transmission, diploid female castes, should be positive for viruses, transmitted in this way. However, we did not detect ABPV, BQCV, CBPV, and SBV in the donor colonies. These results seem to be caused by the absence of the paternal route of vertical virus infection. But finally, we are not able to exclude the paternal route completely, because of the genetically heterogeneity of workers. Queens naturally mate with several drones, while their sperm is stored in the queen spermatheka. Accordingly, we are not able to distinguish between the genetic origin of the sampled workers. In order to exclude the paternal route, single drone-mated queens have to be used (Prodelalova et al., 2019). Horizontal transmission routes of virusesUninfected individuals become infected by contact to infected individuals (Mazzei et al., 2014; Amiri et al., 2020; Yañez et al., 2020). In case of queens during their rearing process, nurse bees are the main risk for horizontal transmission. Within the donor colony, eggs and young larvae come into contact with nurse bees (Seifert et al., 2020, 2021). In the case of infected donor colonies (ABPV, BQCV, CBPV, and SBV), queens might have become infected in their early developmental stages. However, we found no evidence of a correlation between virus detection in donor colonies and queen mortality. Potential horizontal transmission in donor colonies was assumed for three series (IV, VII, and X), because of virus detection in sampled workers. According to Pirk et al. (2013) 5% infected individuals were detected with a probability of 53.67% in this setting. However, a dilution and decrease of infected individuals from series to series could be caused by the natural turnover of bee mass in intact colony structures of donor colonies (Betti et al., 2016). Nurse hives seem to be the main risk for horizontal virus transmission, because of their intense nursing activity on queen larvae. These colonies were sampled weekly during the breeding season. ABPV, BQCV, CBPV, DWV, and SBV were identified in different series. Interestingly, in later series detections ABPV, BQCV, and CBPV seem to be prevalent. Compared to donor colonies, DWV was detected additionally. Only in 23.53% of virus detections, the virus was detected in a single colony, compared to single detections in donor colonies (44.44%). According to the same probability of virus detection in the colony (Pirk et al., 2013), viruses seem to be more prevalent in nurse hives than in donor colonies. Nurse hives do not have a queen, therefore the workers are willing to nurse young larvae to raise a queen. These nurse hives are kept for several weeks in a queen-less status, without any worker or drone brood in larval stages. This unnatural situation is reinforced by adding capped brood frames on a weekly basis. By adding brood frames, viruses as well as phoretic and replicating Varroa mites can be transferred by the beekeeper, leading to an accumulation of virus-positive workers (Ravoet et al., 2015). Whereas queen larvae are the only nursed individuals in these colonies, a higher nursing frequency of virus-positive workers may cause higher viral loads in queen larvae.
Fig. 5. Abundance and quantification of different virus species in queens during the breeding season 2020. Titers of detected virus species were determined (copies/bee). Colored boxplots indicate the cumulated results of series 0–X: green, Aparavirus (ABPV); red, Triatovirus (BQCV); orange, Cripavirus (CBPV); blue, Iflavirus (DWV, SBV, and VDV-1). Abundance of virus species among series 0–X: A, ABPV; B, BQCV; C, CBPV; D, DWV; E, SBV; F, VDV-1. Significance codes: **, 0.01> p > 0.001; ***, p < 0.001.
Fig. 6. Abundance and titer of different virus species, detected in virus-positive queens. Titers of detected virus species were determined (copies/bee). Green, Aparavirus (ABPV); red, Triatovirus (BQCV); orange, Cripavirus (CBPV); blue, Iflavirus (DWV, SBV, and VDV-1). A, relative abundance (%, n=328) of virus species in 2020; B, cumulated titer of detected virus species in 2020. Significance codes: *, 0.05> p > 0.01; ***, p < 0.001.
Fig. 7. Titers of different virus species in female castes of A. mellifera. Titers of different virus species were calculated for female castes of A. mellifera, representing worker of nurse hives and donor colonies, as well as derived queens. Significance codes: *, 0.05> p > 0.01; **, 0.01 < p < 0.001. Finally, the importance of royal jelly needs to be mentioned, since it may be produced by virus-positive workers, and accordingly can horizontally infect nursed queen larvae (Mazzei et al., 2014). Taken together, nursing queen larvae harbor the risks of horizontal and vectorial transmission of several viruses. Vectorial transmission of viruses by V. destructorDirect vertical transmission of viruses is defined in Varroa transmitting viruses during its replication in capped brood cells on pupae. Having a look at donor colonies, a direct vertical transmission on queen eggs and larvae can be excluded while using first instar larvae, which cannot be affected by V. destructor (Zheguang et al., 2018). But vectorial transmission in worker pupae in donor colonies can lead to horizontal infections of queen eggs and larvae in a second step by infected nurse bees. DWV and VDV-1 are closely adapted to V. destructor (Gisder et al., 2009), while both were not detected in donor colonies. Because of the intact social and age structure of donor colonies, as well as the availability of drone brood as trapping combs, fewer worker bees may be affected by V. destructor during the breeding season (Frey and Rosenkranz, 2014; Betti et al., 2016; Zheguang et al., 2018). In contrast to donor colonies, nurse hives represent an unnatural structure without eggs and larva stages of worker and drone brood. Varroa destructor is forced to replicate in bee brood (Rosenkranz et al., 2010). Nursed queen brood is the only source for Varroa replication in nurse hives. Under natural conditions, queen cells are not infested with Varroa mites (Tiesler et al., 2016), because of their intense nursing. However, adding capped brood frames over several weeks leads to an immense dimension of Varroa accumulation in nurse hives, resulting in upcoming Varroa infestation of queen cells in proceeding breeding series. The detection of V. destructor in capped queen cells (data not shown) indicated the direct vertical transmission of viruses on queens. A high Varroa infestation level may also cause a high abundance of DWV in nurse hives (Gisder et al., 2009). According to the presence of different viruses in nurse hives, a horizontal transmission of these viruses by nurse bees was identified as a route of infection. Moreover, these horizontal transmissions can be assumed as a secondary vectorial transmission, as result of vertical infected nurse bees. Queens were analyzed for virus infection individually. The results of virus detection differed from the other sampled bee material regarding the detected virus frequency. BQCV was ubiquitous in all 11 series, without significant deviations within the series. ABPV, CBPV and SBV were not detected in all series, but after their first detection (ABPV: series V; CBPV: series VII; SBV; series III) they did not disappear in the following series. A statistical correlation between queens and their original colonies (donor colonies) and nurse hives could not be established in this study, which might be due to detection probability in donor colonies and nurse hives (Prik et al., 2013). Interestingly, these viruses were detected in series after they were detected in donor colonies and nurse hives. Whereas ABPV was detected in series IV in donor colonies and nurse hives, it was firstly detected in series V in a single queen, followed by further detection in more individuals per series. An analogous appearance was shown for CBPV, which was detected in series VII in queens for the first time; whereas, it was detected in nurse hives already in series IV (donor colonies also in series VII). SBV was detected in series III, but in colonies of the sequential series IV (donor colonies) and VII (nurse hives). This may be explained by the used sample sizes and detection limits in the different bee material. Compared to sequential colonies, two more species of the genus Iflavirus were detected in queens. Both of them, DWV and VDV-1, are closely linked to infestation levels of V. destrucor (Gisder et al., 2009). Because of the nurse hives’ structure, queen larvae strongly attract mites when no larvae of workers and drones are present, while virus-positive adult bees and mites accumulate in the hive. DWV was detected in two series with moderate titers. In contrast, VDV-1 was detected in eight series with obviously increasing titers. VDV-1 is able to replicate in V. destructor, resulting in immense vector-transmitted titers (Gisder and Genersch, 2020), which can lead to the detected amounts of VDV-1. The comparison between virus detection and queen mortality revealed no correlation. Three series showed a significantly reduced mortality and only the last series a significantly increased mortality of queens. Deviations within the series might be explained by abiotic effects, like temperature and especially in the last series by seasonal effects on the honeybees (Tiesler et al., 2016; Beims et al., 2020a, 2020b). Higher mortality was recorded in uncapped queen cells. This observation can be explained by the accessibility of larvae by nurse bees. After capping, nurses do not have direct access to developing queens. Moreover, capped cells were removed from the nurse hive and transferred into an incubator. If queens died after cell capping, they might be recognized as being infected by nurse bees; and therefore, would be removed. When queen pupae die under in vitro conditions in an incubator, their death is recorded at the end of development. Besides the mortality of queens, the detection quote of different viruses was analyzed. A continuous increase in virus detection was observed, starting with approximately 40% in series 0 and 100% in series X. Several virus species were detected with different abundances during the breeding season. DWV and VDV-1 were abundant in low levels from their first detection, but not ubiquitously. These low abundances may be explained by their close relationship to V. destructor, which is rarely able to enter queen cells. But as discussed above, the transfer by the use of nurse hives in a continuous series cannot be excluded. Our results indicate an unproblematic use of nurse hives in the three following series in the case of low initial load with Varroa mites. SBV and ABPV integrated into their detection quote since their first abundance. On the contrary, in DWV variant, their abundance increased in some series, which resulted in local peaks, and decreased afterward. These abundances indicate a relation between infected individuals of nurse bees, which disappeared after their specific span of life outside of the hives (Betti et al., 2016; Tiesler et al., 2016), resulting in lower detection in later series. Regarding the abundances of SBV and VDV-1, it seems that the transfer of infected brood into nurse hives leads to a time shift of infection in nursed queens. CBPV could be detected firstly in series VII with an abundance of approximately 40% and it directly increases its abundances up to 95% within 1 week. This virus seems to spread rapidly, without disappearing according to the life span of the nurse bees (Amiri et al., 2014). BQCV was detected in all series, starting with an abundance of approximately 40%, resulting in 100% positive detections in series X. Explicit deviations could not be observed, but the abundance seems to increase continuously. We assume a persistence of BQCV in nurse colonies, spreading through the nurse hives and affecting more and more queens (Amiri et al., 2020). In contrast to BQCV detection, we did not observe typical BQCV symptoms like black-colored queen cells. Taken together, the titer of ABPV and CBPV differ significantly from other virus species in queens, while the titers of BQCV, DWV, and SBV spread among wide ranges. Perspectives of common sustainable breeding proceduresDuring the rearing and mating of honey bee queens, the risk for transmission of viruses can be reduced. To reduce the risk of maternal vertical virus transmission, a combination of screening the donor queens (2a) successors by SOV (De Graaf et al., 2020) and individual screening is recommended. Further attention should be laid on the paternal route. Therefore, individual screening and SOV in drone colonies (1b) appear to be worthwhile. For higher reliability, this method could be further combined with instrumental insemination of queens. We suggest collecting the drone’s sperm individually. After collecting, the heads of the drones should be used for virus analysis (Schurr et al., 2019). Moreover, samples of e-sperm could be analyzed for viruses (Prodelalova et al., 2019). Together, the combination of these methods could exclude efficiently the transmission of viruses in the vertical route. To minimize the risks of horizontal and vectorial transmission, nurse hives should be used in a single series, to avoid the accumulation of V. destructor. Finally, the breeding process should be performed in restricted areas, like mating stations (Beims et al., 2020a) to decrease the risk of horizontal pathogen transmission (Amiri et al., 2014; Frey and Rosenkranz, 2014; Ravoet et al., 2015; Posda-Florez et al., 2020) between screened and un-screened colonies. Taken together, we detected different viruses in our donor colonies, as well as in the nurse hives. A correlation between virus detection in these colonies and derived queens could not be shown. Moreover, the viral load did not influence the mortality of queens during the rearing process. Our results indicate a lower interrelation between virus abundance in queens and their original donor colonies, than between nurse hives and its nursed queens. However, by preceding the breeding series with artificial queenless nurse hives, the virus abundance in nursed queens increased. Further studies are needed to investigate the potential sublethal effects of virus-infected queens during the rearing process, as well as in their offspring. AcknowledgmentsWe thank Marc Hermann for excellent technical assistance, as well as Stefan Lembke and Paula Markwitz for excellent assistance in beekeeping and providing the colonies material. Conflict of interestThe authors declare that there is no conflict of interest. FundingThis work was funded by the Lower Saxony State Office for Consumer Protection and Food Safety, Institute for Apiculture, Celle, and the Lower Saxony Ministry of Nutrition, Agriculture and Consumer Protection, within project 58-32. Availability of dataThe authors confirm the data generated or analyzed during this study are included in this publication. All data (including raw data) are also available from the corresponding author on reasonable request. Author contributionsConceptualization, B.H. and vdO.W.; methodology, B.H.; software, B.H.; validation, B.H., S.M. and vdO.W.; formal analysis, J.M. and vdO.W.; investigation, B.H.; resources, S.M. and J.M.; data curation, B.H.; writing—original draft preparation, B.H.; writing—review and editing, S.M., vdO.W. and J.M.; visualization, B.H.; supervision, vdO.W.; project administration, B.H.; funding acquisition, vdO.W. All authors have read and agreed to the published version of the manuscript. ReferencesAmiri, E., Meixner, M., Büchler, R. and Kryger, P. 2014. Chronic bee paralysis virus in honeybee queens: evaluatuing susceptibility and infection routes. Viruses 6(3), 1188–1201. Amiri, E., Strand, M.K., Tarpy, D.R. and Ruepell, O. 2020. Honey bee queens and virus infections. Viruses 12(3), 322. Beims, H., Janke, M. and von der Ohe, W. 2020a. Evaluation of drone migration on mating station torfhaus (DE-6-14) in 2020 by cubital index analysis. SSRG. Int. J. Agri. Environ. Sci. 7(5), 38–42. Beims, H., Janke, M., von der Ohe, W. and Steinert, M. 2020b. Rapid identification and genotyping of the honeybee pathogen Paenibacillus larvae by combining culturing and multiplex quantitative PCR. Open. Vet. J. 10(1), 53–58. Betti, M.I., Wahl, L.M. and Zamir, M. 2016. Age structure is critical to the population dynamics and survival of honeybee colonies. R. Soc. Open. Sci. 3(11), 160444. De Graaf, D.C., Laget, D., De Smet, L., Boúúaert, D.C., Brunian, M., Veerkamp, R.F. and Brascamp, E.W. 2020. Heritability estimates of the noveltrait ‘suppressed in ovo virus infection’ in honey bees (Apis mellifera). Sci. Rep. 10(1), 14310. De Miranda, J.R., Bailey, L., Ball, B.V., Blanchard, P., Budge, G., Chejanovsky, N., Chen, Y.P., Gauthier, L., Genersch, E., de Graaf, D., Ribière, M., Ryabov, E., de Smet, L. and van der Steen, J.J.M. 2013. Standard methods for virus research in Apis mellifera. J. Apicult. Res. 52(4), 1–56. De Miranda, J.R., Cordoni, G. and Budge, G. 2010. The acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J. Invertebr. Pathol. 103(Suppl. 1), 30–47. Deutscher Imkerbund. 2017. Richtlinien für das Zuchtwesen des Deutschen Imkerbundes (ZRL). Wachtberg, Germany: Deutscher Imkerbund e.V. Dietemann, V., Pflugfelder, J., Anderson, D., Charriere, J., Chejanivski, N., Daint, B., de Miranda, J., Delaplane, K., Dillier, F.X., Fuchs, S., Gallmann, P., Gauthier, L., Imdorf, A., Koeniger, N., Kralj, J., Meikle, W., Pettis, J., Rosenkranz, P., Sammataro, D., Smith, D., Yanez, O. and Neumann, P. 2012. Varroa destructor: research avenues towards sustainable control. J. Apicult. Res. 51, 125–132. Engel, P., Bartlett, K.D. and Moran, N.A. 2015. The bacterium Frischella perrara causes scrab formation in the gut of its honeybee host. mBio 6(3), e00193–e00115. Frey, E. and Rosenkranz, P. 2014. Autumn invasion rates of Varroa destructor into honey bee colonies and the resulting increase in mite population. J. Econ. Entomol. 107(2), 508–515. Gisder, S., Aumeier, P. and Genersch, E. 2009. Deformed Wing virus: replication and viral load in mites (Varroa destructor). J. Gen. Virol. 90, 463–467. Gisder, S. and Genersch, E. 2015. Special issue: honey bee viruses. Viruses 7(10), 5603–5608. Gisder, S. and Genersch, E. 2020. Direct evidence for infection of Varroa destructor mites with the bee-pathogenic Deformed Wing virus variant B, but not variant A, via fluorescence in situ hybridization analysis. J. Virol; doi:10.1128/JVI.01786-20. Goetze, G.K.L. 1936. Technik und Ergebnisse von Erbversuchen mit der Honigbiene. Deut. Imkerfreund 10, 68–74. Kevill, J.L., Highfield, A., Mordecai, G.J., Martin, S.J. and Schroeder, D.C. 2017. ABC assay: method development and application to quantify the role of three DWV master variants in overwinter colony losses of European honey bees. Viruses 9(11), 314. Mazzei, M., Carrozza, M.L., Luisi, E., Forzan, M., Giusti, M., Sagona, S., Tolari, F. and Felicioli, A. 2014. Infectivity of DWV associated to flower pollen: experimental evidence of a horizontal transmission route. PLoS One 9(11), e113448. McMenamin, A.J. and Flenniken, M.L. 2018. Recently identified bee viruses and their impact on bee pollinator. Curr. Opin. Insect. Sci. 26, 120–129. McMenamin, A.J. and Genersch, E. 2015. Honey bee colony losses and associated viruses. Curr. Opin. Insect. Sci. 8, 121–129. Moore, J., Jironkin, A., Chandler, D., Burroughs, N., Evans, D.J. and Ryabov, E.V. 2011. Recombinants between Deformed wing virus and Varroa destructor virus-1 prevail in Varroa destructor-infested honeybee colonies. J. Gen. Virol. 92, 156–161. Pirk, C.W.W., de Miranda J.R., Kramer, M., Murray, T.E., Nazzi, F., Shutler, D., van der Stehen, J.J.M. and van Dooremalen, C. 2013. Statistical guidelines for Apis mellifera research. J. Apicult. Res. 52(4), 1–24. Posda-Florez, F., Ryabov, E.V., Heerman, M.C., Chen, Y., Evans, J.D., Sonenshine, D.E. and Cook, S.C. 2020. Varroa destructor mites vector and transmit pathogenic honey bee viruses acquired from an artificial diet. PLoS One 15(11), e0242688. Prodelalova, J., Moutelikova, R. and Titera, D. 2019. Multiple virus infections in western honeybee (Apis mellifera L.) ejaculate used for instrumental insemination. Viruses 11(4), 306. Ravoet, J., De Smet, L., Ernseleers, T. and de Graaf, D.C. 2015. Vertical transmission of honey bee viruses in a Belgian queen breeding program. BMC. Vet. Res. 11, 61. Ribière, M., Olivier, V. and Blanchard, P. 2010. Chronic bee paralysis: a disease and a virus like no other? J. Invertebr. Pathol. 103(Suppl. 1), 120–131. Ritten, C.J., Peck, D., Emke, D. and Buddhika-Patalee, M. 2018. Firm efficiency and returns-to-scale in the honey bee pollination service industry. J. Econ. Entomol. 111(3), 1014–1022. Rosenkranz, P., Aumeier, P. and Ziegelmann, B. 2010. Biology and control of Varroa destructor. J. Invertebr. Pathol. 103, 96–119. Ruiz-Villalba, A., van Pelt-Verkuil, E., Gunst, Q.D., Ruijter, J.M. and van den Hoff, M.J.B. 2017. Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR). Biomol. Detect. Quantif. 14, 7–18. Ryabov, E.V., Fannon J.M., Moore, J.D., Wood, G.R. and Evans D.J. 2016. The Iflavirus sacbrood virus and Deformed wing virus evoke different transcriptional responses in the honeybee which may facilitated their horizontal or vertical transmission. Peer. J. 4, e1591. Schurr, R., Tison, A., Militano, L., Cheviron, N., Sircoulomb, F., Rivière, M.P., Ribière-Chabert, M., Thiéry, R. and Dubois, E. 2019. Validation of quantitative real-time RT-PCR assays for the detection of six honeybee viruses. J. Virol. Methods. 270, 70–78. Seifert, P., Buling, N. and Grünewald, B. 2021. Honey bee behaviours within the hive: insights from long-term video analysis. PLoS One 16(3), e0247323. Seifert, P., Hota, R., Ramesh, V. and Grünewald, B. 2020. Chronic within-hive video recordings detect altered nursing behaviour an retarded larval development of neonicotinoid treated honey bees. Sci. Rep. 10, 8727. Spurny, R., Pridal, A., Palkova, L., Kiem, H.K.T., de Miranda, J.R. and Plevka, P. 2017. Virion structure of black queen cell virus, a common honeybee pathogen. J. Virol. 91(6), e02100–e02116. Tantillo, G., Bottaro, M., Di Pinto, A., Martella, V., Di Pinto, P. and Terio, V. 2015. Virus infections of honeybees Apis mellifera. Ital. J. Food. Saf. 4(3), 5364. Tiesler, F.K., Bienefeld, K. and Büchler, R. 2016. Selektion bei der Honigbiene. Herten, Germany: Buschhausen Druck- und Verlagshaus. Traynor, K.S., Mondet, F., de Miranda, J.R., Techer, M., Kowallik, V., Oddie, M.A.Y., Chantawannakul, P. and McAfee, A. 2020. Varroa destructor: a complex parasite, crippling honey bees worldwide. Trends Parasitol. 36(7), 592–606. Van Alphen, J.J.M. and Fernhout, B.J. 2020. Natural selection, selective breeding, and the evolution of resistance of honeybees (Apis mellifera) against Varroa. Zoological. Let. 6, 6. Yañez, O., Piot, N., Dalmon, A., de Miranda, J.R., Chantawannakul, P., Panziera, D., Amiri, E., Smagghe, G., Schroeder, D. and Chejanovsky, N. 2020. Bee viruses: routes of infection in hymenoptera. Front. Microbiol. 11, 943. Zheguang, L., Qin, Y., Page, P., Wang, S., Li, L., Wen, Z., Hu, F., Neumann, P., Zheng, H. and Dietemann, V. 2018. Reproduction of parasitic mites Varroa destructor in original and new honeybee hosts. Ecol. Evol. 8(4), 2135–2145. Ziegelmann, B., Abele, E., Hannus, S., Beitzinger, M., Berg, S. and Rosenkranz, P. 2018. Lithium chloride effectively kills the honey bee parasite Varroa destructor by a systemic mode of action. Sci. Rep. 8(1), 683. | ||
| How to Cite this Article |
| Pubmed Style Beims H, Janke M, Ohe WVD, Steinert M. Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process. Open Vet J. 2023; 13(7): 879-893. doi:10.5455/OVJ.2023.v13.i7.10 Web Style Beims H, Janke M, Ohe WVD, Steinert M. Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process. https://www.openveterinaryjournal.com/?mno=146262 [Access: July 03, 2025]. doi:10.5455/OVJ.2023.v13.i7.10 AMA (American Medical Association) Style Beims H, Janke M, Ohe WVD, Steinert M. Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process. Open Vet J. 2023; 13(7): 879-893. doi:10.5455/OVJ.2023.v13.i7.10 Vancouver/ICMJE Style Beims H, Janke M, Ohe WVD, Steinert M. Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process. Open Vet J. (2023), [cited July 03, 2025]; 13(7): 879-893. doi:10.5455/OVJ.2023.v13.i7.10 Harvard Style Beims, H., Janke, . M., Ohe, . W. V. D. & Steinert, . M. (2023) Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process. Open Vet J, 13 (7), 879-893. doi:10.5455/OVJ.2023.v13.i7.10 Turabian Style Beims, Hannes, Martina Janke, Werner Von Der Ohe, and Michael Steinert. 2023. Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process. Open Veterinary Journal, 13 (7), 879-893. doi:10.5455/OVJ.2023.v13.i7.10 Chicago Style Beims, Hannes, Martina Janke, Werner Von Der Ohe, and Michael Steinert. "Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process." Open Veterinary Journal 13 (2023), 879-893. doi:10.5455/OVJ.2023.v13.i7.10 MLA (The Modern Language Association) Style Beims, Hannes, Martina Janke, Werner Von Der Ohe, and Michael Steinert. "Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process." Open Veterinary Journal 13.7 (2023), 879-893. Print. doi:10.5455/OVJ.2023.v13.i7.10 APA (American Psychological Association) Style Beims, H., Janke, . M., Ohe, . W. V. D. & Steinert, . M. (2023) Influence of virus abundances in donor colonies and nurse hives on queens of Apis mellifera during the rearing process. Open Veterinary Journal, 13 (7), 879-893. doi:10.5455/OVJ.2023.v13.i7.10 |