| Original Article | ||
Open Vet J. 2020; 10(3): 347-353 doi: 10.4314/ovj.v10i3.14 Open Veterinary Journal, (2020), Vol. 10(3): 347–353 Original Research http://dx.doi.org/10.4314/ovj.v10i3.14 Detection of an emerging novel sublineage Ind2001BD1 and lineage PanAsia of foot-and-mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018Md Zulfekar Ali1* and Md Giasuddin1,21Animal Health Research Division, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh 2Research on FMD and PPR in Bangladesh, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh *Corresponding Author: Md Zulfekar Ali. Animal Health Research Division, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh. Email: zulfekar [at] blri.gov.bd Submitted: 25/04/2020 Accepted: 21/09/2020 Published: 30/09/2020 © 2020 Open Veterinary Journal
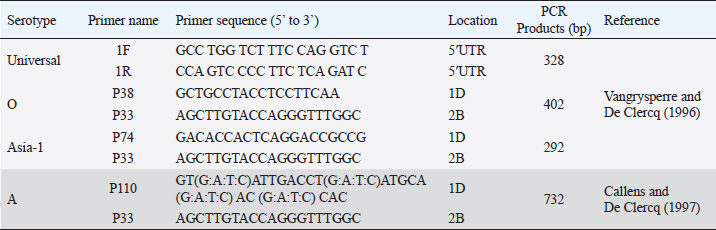
AbstractBackground: Foot-and-mouth disease (FMD) is an endemic disease of cloven-hoofed animals in Bangladesh and multiple outbreaks occur every year because of the FMD virus (FMDV). Aim: The aim of the present investigation was to determine the molecular characterization of the VP1 coding region of FMDV serotype O outbreak in cattle. Methods: A total of four tongue epithelial specimens were collected from clinically FMD-positive cattle during June 2018 in Manikgonj district of Bangladesh. Results: All four isolates were recorded positive for FMDV serotype O. The phylogenetic analysis showed that two isolates were clustered within an emerging novel sublineage Ind2001BD1 under lineage Ind2001 of FMDV serotype O, which was identified during 2012–2016 in Bangladesh. One isolate was clustered within the lineage PanAsia of FMDV serotype O and was closely related to an isolate identified in Nepal in 2009. The phylogenetic reconstruction revealed that all the four isolates belong to the Middle East–South Asia topotype. Conclusion: Therefore, multiple lineages of the FMDV serotype O are circulating among the cattle in the outbreak area, which make it more complex for the FMD control program in Bangladesh. A comprehensive study on the genetic characteristics of FMDV across the country is required for effective FMD prevention and control strategy. Keywords: Cattle, Foot-and-mouth disease, Ind2001BD1, Lineage, PanAsia. IntroductionFoot-and-mouth disease (FMD) is a viral disease caused by the FMD virus (FMDV), a member of the Aphthovirus genus of the Picornaviridae family (Bachrach, 1968; Domingo et al., 2002). It is an acute, highly contagious disease among cloven-hoofed animals, like cattle, buffalo, pigs, sheep, goats, wild ruminants, etc. The prominent clinical signs of FMD are high fever, drooling of saliva, vesicles on tongue, gums, and interdigital spaces (Kitching, 2002; Grubman and Baxt, 2004). Therefore, the animal becomes emaciated, resulting in loss of meat and milk production, abortion of pregnant cows, death of infected young calves, restriction in international trade, and medication cost of infected animals (Grubman and Baxt, 2004; Brito et al., 2017). In Bangladesh, FMD is well known among the village people by its local name “Khura Rog” (Kazi et al., 2007; Bary et al., 2018). In the outbreak areas, an overall of 53.89% cases was reported, among which case fatality rates were 2.27% in adults but very high (71.46%) in calves aged below 2 years in Bangladesh (Ali et al., 2019). The virus can transmit via air and has a transboundary potentiality by the movement of livestock across borders (Loth et al., 2011). In South Asia, there is a record of extensive cattle movement from India, Nepal, and Myanmar to Bangladesh as a neighboring country (Sumption et al., 2008; Khounsy et al., 2009). However, the movement of cattle increases during the Muslim celebration of Eid al-Adha for cattle sacrifice (Gleeson, 2002; Siddique et al., 2018). There are seven serotypes of FMDV, namely serotype A, O, C, Asia 1, SAT 1, SAT 2, and SAT 3 distributed globally. Out of these, only three serotypes A, O, and Asia 1 are circulating in Bangladesh, with serotype O dominating the others (Nandi et al., 2015; Ullah et al., 2015; Ali et al., 2016). But there is no cross-protection between the serotypes and vaccines, and so multivalent strain vaccine is a popular solution for FMD vaccine (Mumford, 2007). Therefore, the genetic characteristics and serotype variations should be taken into consideration for vaccination strategy or any intervention policies in endemic countries (Loth et al., 2011; Siddique et al., 2018). The FMDV has a single-stranded positive-sense RNA genome and is surrounded by 60 copies of icosahedral capsid; each copy is composed of four structural proteins, including VP1, VP2, VP3, and VP4 (Carroll et al., 1984; Jackson et al., 2003). Based on VP1 coding nucleotide sequences, the topotype of FMDV was classified (Samuel and Knowles, 2001). In the Indian subcontinent, the Middle East–South Asia (ME–SA) topotype is circulating and four distinct lineages have been established, namely Ind2001, Ind2011, PanAsia, and PanAsia2, under this topotype (Subramaniam et al., 2015; Brito et al., 2017). Since 2009, lineage Ind2001 of the FMDV serotype O has been dominating in Bangladesh and two novel sublineages Ind2001BD1 and Ind2001BD2 have emerged from 2012 to 2016 (Siddique et al., 2018). Therefore, this study is focused on the investigation of FMDV serotypes and their genetic characteristics from an outbreak visit during 2018. Materials and MethodsCollection of samplesAn FMD outbreak field visit was conducted during June 2018 in four villages of Manikgonj district of Bangladesh. Tongue epithelial specimens were collected from the affected cattle of each village into a virus transfer media. The geographic positions (23.905632, 89.956403; 23.901865, 89.981122; 23.890565, 89.996228; and 23.886798, 90.011334) of the sampling locations were recorded by GPS mobile application and a sampling map (Fig. 1) was designed by using ESRI ArcGIS version 10.6.1 (ESRI ArcGIS for Desktop, 2018). After collection, the samples were kept in an ice-cool carrier and transferred to FMD Research Laboratory, Bangladesh Livestock Research Institute, and stored at −70°C until laboratory analysis. Detection of FMDV serotypes by RT-PCR testThe tongue epithelial tissue specimens were ground with a pestle and mortar in phosphate-buffered saline solution using sterile sand to make 10% suspension. The genomic total RNA was extracted from an aliquot of suspension samples by using QIAamp® Viral RNA kit (QIAGEN, Hilden, Germany), following the manufacturer’s protocol. The RNAs were subjected to RT-PCR test targeting 5′ UTR and 3D regions for the detection of FMDV, serotypes A, O, and Asia 1 by using QIAGEN One-Step RT-PCR Kit (QIAGEN, Hilden, Germany) with the universal primers listed in Table 1. The suspensions of samples were inoculated into BHK21 cells and incubated for virus isolation. Table 1. List of primer sequences used for universal FMDV and serotyping of FMDV by RT-PCR
Sequencing of VP1 coding regionThe cell culture supernatant, at 3rd passages, was harvested and total RNAs were extracted by using the QIAamp® Viral RNA kit (QIAGEN, Hilden, Germany), following the manufacturer’s protocol. The RNAs were subjected to amplify the VP1 region using the QIAGEN One-Step RT-PCR Kit (QIAGEN, Hilden, Germany) with three specific sets of forward and one reverse primer developed by Knowles et al. (2016) for FMD serotype O. Then, the amplified product was run in 1.5% agarose gel along with 1kb DNA ladder stained with SYBR Safe DNA Gel Stain (ThermoFissher, Waltham, MA) and the amplified band was visible through the UV transilluminator. The amplified product was harvested and extracted by using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) and the product was quantified by using a spectrophotometer (Thermo Scientific™ NanoDrop™, Wilmington DE). The cycle sequencing reactions were carried out on amplified products from a commercial source (Macrogen®, South Korea). Phylogenetic analysisA total of 50 FMDV serotype O sequences were retrieved from the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) along with the four sequences generated in the study to compare the Bangladeshi isolates with the spatially, temporally matching FMDV serotype O circulating in neighboring countries (India and Nepal). The topology, lineage, sublineage, and clade were also carried out during virus collection. The nucleotide sequences were aligned by using BioEdit 7.2.5 (Hall, 1999) and Clustal W 1.83 (Thompson et al., 1994) software. An evolutionary analysis of viruses was conducted in MEGA X (Kumar et al., 2018) using the Maximum Likelihood phylogenetic tree with Tamura–Nei’s model (Tamura and Nei, 1993). The 1,000 times robustness of the branches of the phylogenetic tree was generated to verify the statistical significance.
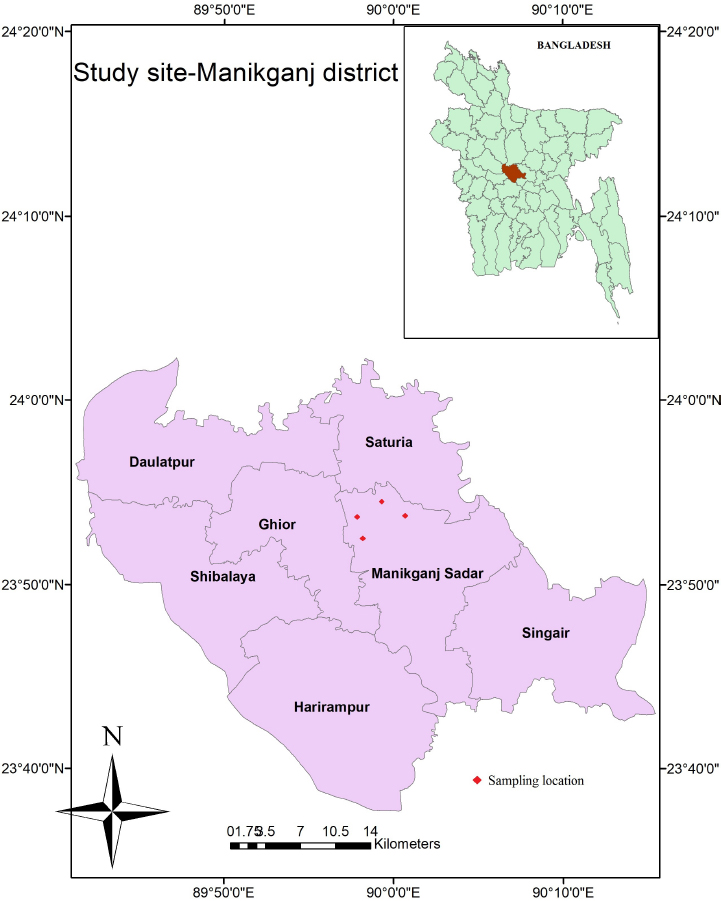
Fig. 1. Map showing the geographical positions of the four samples collected from Manikgonj district of Bangladesh. The map was created by using the Geographic Information Systems (GIS) software ESRI ArcGIS version 10.6.1.
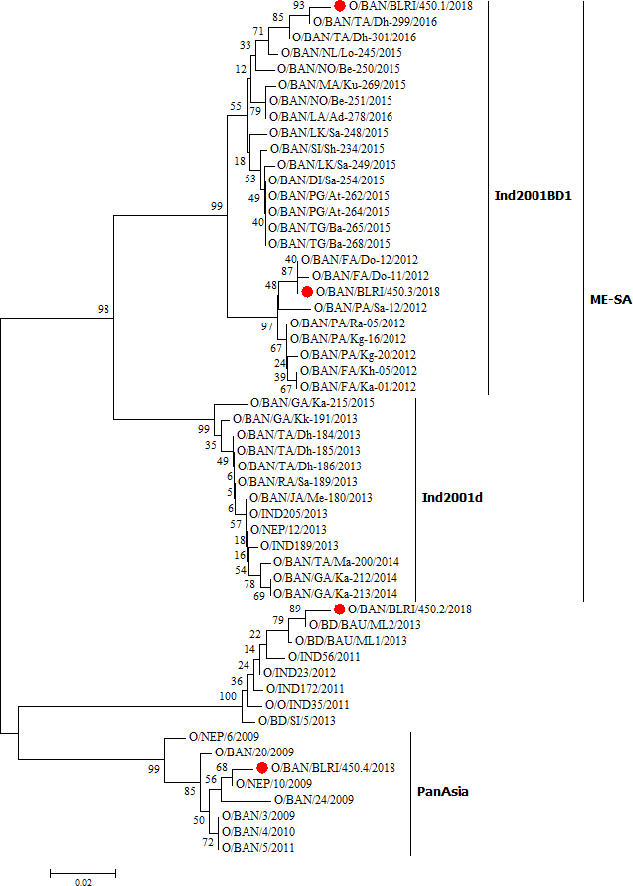
Fig. 2. The phylogenetic analysis of collected isolates and FMDV serotype O from Bangladesh, India, and Nepal from 2009 to 2016. The nucleotide sequences of the VP1 coding region constructed the phylogenetic tree by using the Tamura–Nei model under the Maximum Likelihood method. The nucleotide sequences that were generated for this study are shown in red circles. Ethical approvalThe research protocol was approved by the Animal Experimentation Ethics Committee, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh, under reference-BLRI0006. ResultsFMDV serotype O was detected in all the four samples by RT-PCR test and the virus was isolated in BHK21 cell culture. The VP1 coding region of four isolates was sequenced and submitted to the NCBI GenBank database under assigned accession numbers MT316587–MT316590. The phylogenetic analysis revealed that all the four isolates of FMDV serotype O belong to the ME-SA topotype. Two isolates, including O/BAN/BLRI/450.1/2018 and O/BAN/BLRI/450.3/2018, became clustered within the sublineage Ind2001BD1, a newly emerged novel sublineage of lineage Ind2001 under ME-SA topotype identified in Bangladesh during 2012–2016 (Fig. 2). One isolate was clustered within the PanAsia lineage, including O/BAN/BLRI/450.4/2018. Another one isolate O/BAN/BLRI/450.2/2018 was clustered in an unnamed lineage. Analysis of sequences showed that the isolates of FMDV serotype O from Bangladesh were in a very close relationship with all sequences of FMDV serotype O in Bangladesh and the isolates of FMDV serotype O from Nepal in 2009 (O/BAN/BLRI/450.4/2018, less than 1.5% difference with O/NEP/10/2009). DiscussionFMD is an endemic disease among cattle and buffalos in Bangladesh for many years (WAHID, 2009; Loth et al., 2011). There are three serotypes of FMDV that are circulating in Bangladesh, namely serotype A, O, and Asia 1, with a prevalence of 10%–15%, 80%–85%, and 5%, respectively (Nandi et al., 2015). Therefore, FMDV serotype O is the dominating serotype over the other two serotypes (Giasuddin et al., 2016; Siddique et al., 2018). Every year a lot of FMD outbreak waves occur among the cattle of Bangladesh, which in turn causes an important economic loss. The global impact of FMD is US$ 6.5–21 billion for endemic countries and US$ >1.5 billion for FMD-free countries (Knight-Jones and Rushton, 2013). Howlader et al. (2004) estimated an economic loss of US$ 163,329 for 3,708 affected farms in Bangladesh due to FMD. However, the total economic loss due to FMD is US$ 60 million per year in Bangladesh (FAO/OIE, 2012). Since 2009, lineage Ind2001 under the ME-SA topotype of FMDV serotype O is dominating most of the outbreaks in Bangladesh (Loth et al., 2011; Siddique et al., 2018). In this study, two isolates were clustered with the sublineage Ind2001BD1 of lineage Ind2001 under the ME-SA topotype. The sublineages Ind2001BD1 and Ind2001BD2 are the two novel emergences of the FMDV serotype O in Bangladesh identified by Siddique et al. (2018). They identified 23 isolates under Ind2001BD1 and three isolates under Ind2001BD2 sublineage collected during 2012–2016 in Bangladesh. The genetic analysis showed that two isolates were closely related to isolates collected in 2012 and 2016 by Siddique et al. (2018). So, the novel sublineage Ind2001BD1 was well circulating in the Manikgonj district of Bangladesh where outbreaks occurred. The lineage Ind2001 has been divided into four separate sublineages: Ind2001a, Ind2001b, Ind2001c, and Ind2001d (Subramaniam et al., 2015). Among the sublineages, sublineage Ind2001d is the second most circulating and dominating in Bangladesh from 2009 (Loth et al., 2011; Siddique et al., 2018). But sublineage Ind2001d was not identified in this investigation, which could be a reason for the small sample size, short geographic area, and similar host. The study revealed one more isolate O/BAN/BLRI/450.4/2018 clustered within lineage PanAsia and closely related to FMDV serotype O isolated in late 2009 in Nepal (O/NEP/10/2009), with a difference of less than 1.5%. This possibly indicates a linkage of viruses between the two countries (Loth et al., 2011). A study was conducted by Giasuddin et al. (2016) who reported that the PanAsia lineage of FMDV serotype O was circulating in Bangladesh during 2011–2014. The other research group also reported that the PanAsia lineage of FMD serotype O was circulating in Bangladesh during 2012–2016 (Siddique et al., 2018). In the Indian subcontinent, there are four distinct lineages, namely Ind2001, PanAsia, PanAsia-2, and Ind2011, of the FMDV serotype O which were circulating (Subramaniam et al., 2013). The cattle trade network among India, Myanmar, and Nepal through the international border is very common and increases during big Muslim religious celebrations in Bangladesh (Loth et al., 2011). So, there is a big chance of transboundary movement of viruses from neighboring countries to Bangladesh, which is a main reason for implementing a successful FMD control program in Bangladesh. ConclusionThe investigation revealed an emerging novel sublineage Ind2001BD1 within lineage Ind2001 and lineage PanAsia along with one unnamed cluster of FMDV serotype O circulating in the outbreak area of Bangladesh. There was a closely related FMDV serotype O with a virus of Nepal, which was identified and could be a transboundary linkage. However, comprehensive studies on the current circulating viruses along with proper outbreak investigations are needed to know the genetic characteristics of viruses in Bangladesh for helping further policy decisions of FMD prevention strategy. AcknowledgmentsThis research study was funded by the development project of ‘Research on FMD and PPR in Bangladesh’ (Award no. MOFL-7080) of the Ministry of Fisheries and Livestock, Bangladesh. The authors would like to thank M.A. Sabur and Eusha Islam, members of the outbreak investigation team. The authors are also thankful to Shariful Islam for designing the sampling map. Authors’ contributionM.Z.A. designed the study, collected the sample, conduced the laboratory analysis, interpreted results, analyzed the data, and wrote the manuscript. M.G conceptualized and designed the study, reviewed the manuscript, and supervised the study. ReferencesAli, M.R., Ullah, H., Siddique, M.A., Sultana, M. and Hossain, M.A. 2016. Complete genome sequence of pig-originated foot-and-mouth disease virus serotype O from Bangladesh. Genome Announc. 4(5), e01150-16. Ali, M.Z., Islam, E. and Giasuddin, M. 2019. Outbreak investigation, molecular detection, and characterization of foot and mouth disease virus in the Southern part of Bangladesh. J. Adv. Vet. Anim. Res. 6(3), 346–354. Bachrach, H.L. 1968. Foot-and-mouth disease. Annu. Rev. Microbiol. 22(1), 201–244. Bary, M.A., Ali, M.Z., Chowdhury, S., Mannan, A., Nur e Azam, M., Moula, M.M., Bhuiyan, Z.A., Shaon, M.T.W. and Hossain, M.A. 2018. Prevalence and molecular identification of haemoprotozoan diseases of cattle in Bangladesh. Adv. Anim. Vet. Sci. 6(4), 176–182. Brito, B.P., Rodriguez, L.L., Hammond, J.M., Pinto, J. and Perez, A.M. 2017. Review of the global distribution of foot-and-mouth disease virus from 2007 to 2014. Transbound. Emerg. Dis. 64(2), 316–332. Callens, M. and De Clercq, K. 1997. Differentiation of the seven serotypes of foot-and-mouth disease virus by reverse transcriptase polymerase chain reaction. J. Virol. Methods 67(1), 35–44. Carroll, A.R., Rowlands, D.J. and Clarke, B.E. 1984. The complete nucleotide sequence of the RNA coding for the primary translation product of foot and mouth disease virus. Nucleic Acids Res. 12(5), 2461–2472. Domingo, E., Baranowski, E., Escarmís, C. and Sobrino, F. 2002. Foot-and-mouth disease virus. Com. Immunol. Microb. Infect. Dis. 25(5–6), 297–308. ESRI ArcGIS for Desktop. 2018. Geographic Information Systems (GIS) software ESRI ArcGIS105 version 10.2.1, 2018. Available via http://www.esri.com/software/arcgis/arcgis-for-desktop (Accessed 11 April 2020). FAO/OIE. 2012. FMD virus pools and the regional programmes Virus Pool 2-South Asia. FAO/OIE Global Conference on foot and mouth disease control. Bangkok, Thailand, 27-29 June 2012. Giasuddin, M., Mahmud, M.S., Alam, S.M.S., Samad, M.A., Islam, M.R., Ahasan, M.D., Rahman, M.H., Karim, M.R. and Acharjee, P. 2016. Molecular epidemiology of foot-and-mouth disease viruses circulated in Bangladesh from 2011–2014. Br. Microbiol. Res. J. 16(4), 1–13. Gleeson, L.J. 2002. A review of the status of foot and mouth disease in South-East Asia and approaches to control and eradication. Rev. Sci. Tech. 21(3), 465–472. Grubman, M.J. and Baxt, B. 2004. Foot-and-mouth disease. Clin. Microbiol. Rev. 17(2), 465–493. Hall , T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. InNucleic acids symposium series 1999 Jan 1. [London]: Information Retrieval Ltd., c1979-c2000. 41(41), 95–98. Howlader, M.M., Mahbub-E-Elahi, A.T. and Coll, S.G. 2004. Foot and mouth disease in Baghabari milk shed area and it’s economic loss in Bangladesh. J. Biol. Sci. 4(5), 581–583. Jackson, T., King, A.M., Stuart, D.I. and Fry, E. 2003. Structure and receptor binding. Virus Res. 91(1), 33–46. Kazi, M.K., Basant, S., Sharma, M.C., Ratala, D.R. and Akram, M. 2007. Parasitic diseases. Handbook on livestock and poultry diseases in SAARC countries, Dhaka, Bangladesh: SAARC AC, pp: 124–159. Khounsy, S., Conlan, J.V., Gleeson, L.J., Westbury, H.A., Colling, A., Paton, D.J., Ferris, N.P., Valarcher, J.F., Wadsworth, J., Knowles, N.J. and Blacksell, S.D. 2009. Molecular epidemiology of foot-and-mouth disease viruses from South East Asia 1998–2006: the Lao perspective. Vet. Microbiol. 137(1–2), 178–183. Kitching, R.P. 2002. Clinical variation in foot and mouth disease: cattle. Rev. Sci. Tech. 21(3), 499–502. Knight-Jones, T.J. and Rushton, J. 2013. The economic impacts of foot and mouth disease–What are they, how big are they and where do they occur? Prev. Vet. Med. 112(3–4), 161–173. Knowles, N.J., Wadsworth, J., Bachanek-Bankowska, K. and King, D.P. 2016. VP1 sequencing protocol for foot and mouth disease virus molecular epidemiology. Rev. Sci. Tech. 35(3), 741–755. Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35(6), 1547–1549. Loth, L., Osmani, M.G., Kalam, M.A., Chakraborty, R.K., Wadsworth, J., Knowles, N.J., Hammond, J.M. and Benigno, C. 2011. Molecular characterization of foot-and-mouth disease virus: implications for disease control in Bangladesh. Transbound. Emerg. Dis. 58(3), 240–246. Mumford, J.A. 2007. Vaccines and viral antigenic diversity. Rev. Sci. Tech. 26(1), 69. Nandi, S.P., Rahman, M.Z., Momtaz, S., Sultana, M. and Hossain, M.A. 2015. Emergence and distribution of foot-and-mouth disease virus serotype a and O in Bangladesh. Transbound. Emerg. Dis. 62(3), 328–331. Samuel, A.R. and Knowles, N.J. 2001. Foot-and-mouth disease type O viruses exhibit genetically and geographically distinct evolutionary lineages (topotypes). J. Gen. Virol. 82(3), 609–621. Siddique, M.A., Ali, M.R., Alam, A.S.M.R.U., Ullah, H., Rahman, A., Chakrabarty, R.P., Amin, M.A., Hoque, S.A., Nandi, S.P., Sultana, M. and Hossain, M.A. 2018. Emergence of two novel sublineages Ind2001 BD 1 and Ind2001 BD 2 of foot-and-mouth disease virus serotype O in Bangladesh. Transbound. Emerg. Dis. 65(4), 1009–1023. Subramaniam, S., Mohapatra, J.K., Das, B., Sanyal, A. and Pattnaik, B. 2015. Genetic and antigenic analysis of foot-and-mouth disease virus serotype O responsible for outbreaks in India during 2013. Infect. Genet. Evol. 30, 59–64. Subramaniam, S., Pattnaik, B., Sanyal, A., Mohapatra, J.K., Pawar, S.S., Sharma, G.K., Das, B. and Dash, B.B. 2013. Status of Foot-and-mouth Disease in India. Transbound. Emerg. Dis. 60(3), 197–203. Sumption, K., Rweyemamu, M. and Wint, W. 2008. Incidence and distribution of foot-and-mouth disease in Asia, Africa and South America; combining expert opinion, official disease information and livestock populations to assist risk assessment. Transbound. Emerg. Dis. 55(1), 5–13. Tamura, K. and Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10(3), 512–526. Thompson, J.D., Higgins, D.G. and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22), 4673–4680. Ullah, H., Siddique, M.A., Al Amin, M., Das, B.C., Sultana, M. and Hossain, M.A. 2015. Re-emergence of circulatory foot-and-mouth disease virus serotypes Asia1 in Bangladesh and VP1 protein heterogeneity with vaccine strain IND 63/72. Lett. Appl. Microbiol. 60(2), 168–173. Vangrysperre, W. and De Clercq, K. 1996. Rapid and sensitive polymerase chain reaction based detection and typing of foot-and-mouth disease virus in clinical samples and cell culture isolates, combined with a simultaneous differentiation with other genomically and/or symptomatically related viruses. Arch. Virol. 141(2), 331–344. WAHID. 2009. OIE World Animal Health Database. Available via https://www.oie.int/wahis_2/public/wahid.php/Wahidhome/Home (Accessed 11 April 2020). | ||
| How to Cite this Article |
| Pubmed Style Ali MZ, Giasuddin M. Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018. Open Vet J. 2020; 10(3): 347-353. doi:10.4314/ovj.v10i3.14 Web Style Ali MZ, Giasuddin M. Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018. https://www.openveterinaryjournal.com/?mno=101888 [Access: July 06, 2025]. doi:10.4314/ovj.v10i3.14 AMA (American Medical Association) Style Ali MZ, Giasuddin M. Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018. Open Vet J. 2020; 10(3): 347-353. doi:10.4314/ovj.v10i3.14 Vancouver/ICMJE Style Ali MZ, Giasuddin M. Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018. Open Vet J. (2020), [cited July 06, 2025]; 10(3): 347-353. doi:10.4314/ovj.v10i3.14 Harvard Style Ali, M. Z. & Giasuddin, . M. (2020) Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018. Open Vet J, 10 (3), 347-353. doi:10.4314/ovj.v10i3.14 Turabian Style Ali, Md Zulfekar, and Md Giasuddin. 2020. Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018. Open Veterinary Journal, 10 (3), 347-353. doi:10.4314/ovj.v10i3.14 Chicago Style Ali, Md Zulfekar, and Md Giasuddin. "Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018." Open Veterinary Journal 10 (2020), 347-353. doi:10.4314/ovj.v10i3.14 MLA (The Modern Language Association) Style Ali, Md Zulfekar, and Md Giasuddin. "Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018." Open Veterinary Journal 10.3 (2020), 347-353. Print. doi:10.4314/ovj.v10i3.14 APA (American Psychological Association) Style Ali, M. Z. & Giasuddin, . M. (2020) Detection of emerging novel sublineage Ind2001BD1 and lineage PanAsia of Foot-and mouth disease virus serotype O in cattle in Manikgonj district of Bangladesh, 2018. Open Veterinary Journal, 10 (3), 347-353. doi:10.4314/ovj.v10i3.14 |