| Research Article | ||
Open Vet. J.. 2025; 15(3): 1178-1186 Open Veterinary Journal, (2025), Vol. 15(3): 1178-1186 Research Article Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in BaghdadMoutaz A.W. Abdul Mounam1, Basil R.F. Razook2 and Entesar Hussain Madi2*1Department of Veterinary Public Health, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq 2Department of Zoonoses Research Unit, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq *Corresponding Author: Entesar Hussain Madi. Department of Zoonoses Research Unit, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq. Email: Intisar.h [at] covm.uobaghdad.edu.iq Submitted: 13/11/2024 Accepted: 16/02/2025 Published: 31/03/2025 © 2025 Open Veterinary Journal
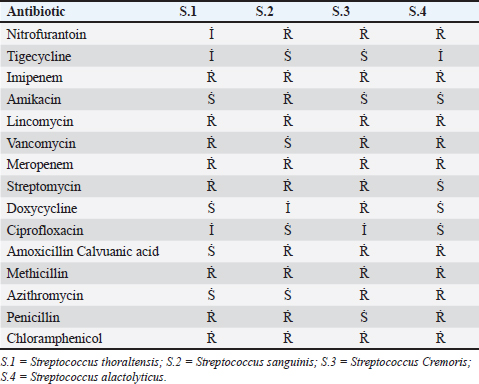
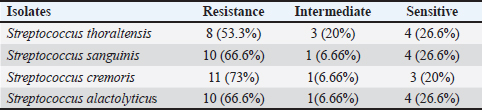
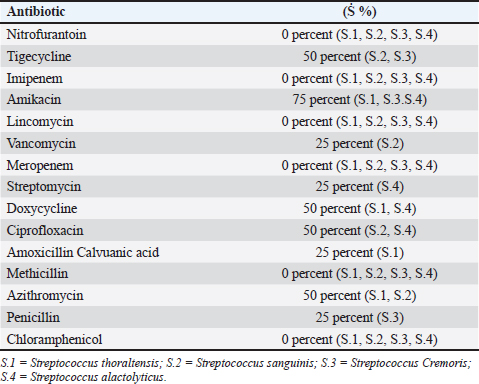
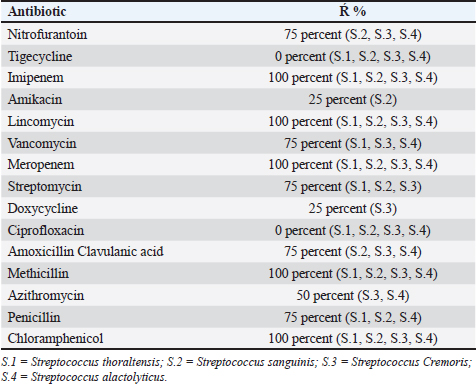
AbstractBackground: This research identified Streptococci spp. depending on culture, biochemistry, the Visual Information Technologies (VITEK) technique, ability to produce biofilms, and antibiotic resistance. Aim: The goal of this study was to perform microbiological procedures to evaluate the qualitative qualities of mozzarella cheese against infective Streptococci using microbiological care. Methods: Sixty (60) mozzarella cheese samples were brought from diverse markets in Baghdad from October 2023 to December 2023 at the Zoonoses Research Unit and Veterinary Public Health Department, Veterinary Medicine College, University of Baghdad. Culture of samples on agar (MacConkey and blood) and aerobically incubated at 37°C for 48 hours. Gram staining purified colonies to notice Gram-positive cocci arranged in strings that were supposed to be Streptococcus species, and then sub cultured on nutrient agar for identification using the VITEK technique. The isolates were tested for their capacity to lyse human red blood cells by streaking blood agar and incubating at 37°C for 24 hours. Streptococcal isolates were streaked on a Congo-red medium. All isolates were inspected for antibiotic susceptibility using 15 common antibiotics via the disc diffusion method. Results: Isolates mozzarella cheese samples 8 (13.3%), these (eight) isolates include: (Streptococcus thoraltensis, Streptococcus sanguinis, Streptococcus cremoris, and Streptococcus alactolyticus. The susceptibility of S. thoraltensis, S. sanguinis, and S. alactolyticus was (26.6%), and S. cremoris (20%) to all antibiotics. Resistance of S. thoraltensis was (53%), S. sanguinis, S. alactolyticus were (66%), and S. cremoris was (73%) to all antibiotics. S. thoraltensis, S. cremoris, and S. alactolyticus were susceptible (75%) to Amikacin. S.sanguinis and S. cremoris were susceptible (50%) to Tigecycline. Streptococcus thoraltensis and S. alactolyticus were susceptible (50%) to Doxycycline. Streptococcus sanguinis and S. alactolyticus were susceptible (50%) to Ciprofloxacin. Streptococcus thoraltensis and S. sanguinis were susceptible (50%) to Azithromycin. Streptococcus sanguinis was susceptible (25%) to vancomycin. Streptococcus alactolyticus was susceptible (25%) to Streptomycin. Streptococcus thoraltensis was susceptible (25%) to Amoxicillin and Clavulanic acid. S. cremoris was susceptible (25%) to Penicillin. All isolates were 100% resistant to imipenem, lincomycin, meropenem, methicillin, and chloramphenicol. Conclusion: Analysis of mozzarella cheese samples identified four predominant Streptococcus species and their antibiotic activity. Keywords: Streptococci isolates, Streptococcus spp., Antibiotics susceptible, Mozzarella cheese, Unpasteurized milk. IntroductionThe genus Streptococcus contains both commensal and harmful species. It has experienced tremendous extension and revision as a result of the speedy development of technologies for molecular identification and microbial phenotyping (Lu et al., 2016; Tian et al 2019). Streptococcus has about 100 recognized species, numerous of which are nurses or symbiodinium in people and animals (Foster et al., 2020). Antibiotic resistance is transformed from beneficial and commensal bacteria to pathogenic bacteria through the docximportant role of the food chain (Flórez and Mayo, 2017). Streptococci are widely spread in humans and animals, and they can cause various diseases (Dhanda et al., 2013). They are present in the oral and intestinal regions of animals and humans, as well as in raw milk, dairy products, and plant material (Tsakalidou et al., 1998). The source of the milk (sheep or cows), method of production (unpasteurized milk and pasteurized or aging period), production, conditions, and other factors affect the microorganisms in the cheese (Verraes et al., 2014; Yao et al., 2022). Dairy products, due to their short shelf life, serve as an ideal growth medium for a diverse array of microorganisms (Ledenbach and Marshall, 2009; Garnier et al., 2017; Rauh and Xiao, 2022). Therefore, it is important to monitor the microbiological quality of dairy (products), especially the whole viable count and concentration of Escherichia coli bacteria, as they are indicators of the sanitary condition of these (products) (Ryu et al., 2012; Pal et al., 2016). The concentration of lactic acid bacteria (LAB) in dairy products such as mozzarella cheese, which are the main components of the starter culture, should be monitored (Mazzoli et al., 2014; Blaya et al., 2018). Fermented products enhance the flavor and consistency of cheese while preventing food spoilage bacteria by producing substances that hinder their growth (Najim, 2010; Algboory et al., 2017; Hussein and Jabbar, 2020). Cheese makers must frequently observe the concentrations of LAB and bacteria during the production of fresh cheese to ensure the quality and safety of their products (Losito et al., 2014). Antimicrobial resistance is a global problem (Watkins and Bonomo, 2016; Motaweq and Naher, 2017; Zhang et al., 2019). Antibiotic-resistant bacteria have been identified in various foods (Wang et al., 2012; De Jong et al., 2013). Devirgiliis et al. (2013), Gad et al. (2014), and Soares-Santos et al. (2015) detailed the factors that influence antibiotic resistance in bacteria associated with dairy products. They also highlighted the widespread distribution of antibiotic resistance genes among food bacteria, emphasizing their role as reservoirs of resistance genes (Marshall and Levy, 2011; Hu et al., 2013; Flayyih et al., 2016; Albanna and Al-Layla, 2020; Taher et al., 2020). In a study, infection with Streptococcus isolated from soft mozzarella cheese, and the importance of stopping the spreading of the epidemic by using molecular tools, first by detecting pathogenic microbes in food, which pose a health hazard to consumers (Hussaini et al., 2014; Mounam et al., 2023). Materials and MethodsThe research was conducted from October 2023 to December 2023 at the Zoonotic Disease Unit and the Public Health Department of the Veterinary Medicine College, University of Baghdad. Sixty (60) mozzarella cheese samples were emulsified with 2% buffered sodium citrate in a stomacher for 3 minutes and subsequently cultured on (MacConkey agar and blood agar). The cultures were then incubated aerobically at 37°C for 48 hours as described by Coque et al. (1995). The purified colonies were subjected to Gram staining, which revealed cocci arranged in series, suggesting Streptococcus species. These colonies were further subcultured on nutrient agar and were identified using the VITEK method. VITEK methodThe VITEK technique was performed following the manufacturer’s instructions. The isolated colonies were moved into a polystyrene tube filled with a saline solution (NaCl) adjusted to a density range of 0.5–0.63. The tube and card were placed in the VITEK 2 cassette using the VITEK 2 Densi Check spectrophotometer. The cassette was then autoincubated inside the VITEK two instrument, and results were interpreted after 18–24 hours. This procedure was performed at the Zoonoses Unit laboratories of the Veterinary Medicine College, University of Baghdad. Hemolysin activityHemolysis is a critical virulence factor in Streptococcus infections, influencing infection severity. Streptococcal isolates were evaluated for their capacity to lyse red blood cells, streak the colonies on blood agar, and incubate them at 37°C for 24 hours. The appearance of a clear zone (halo) around the colonies indicated hemolysin activity, and the type of hemolysis was recorded (Coque et al., 1995). Biofilm formationBiofilm formation was evaluated by streaking Streptococcal isolates on Congo-red medium and incubating them at 37°C for 24 hours (Fig. 1). Following the method outlined by Freeman et al. (1989). Antimicrobial susceptibility testThe Kirby–Bauer method uses paper disks infused with antibiotics placed on the surface of Muller–Hinton agar to assess antibiotic susceptibility. After incubating 4–5 colonies in Tryptone soy broth at 37°C until reaching a standard opacity (equal to a 0.5 McFarland standard), a sterile swab was used to spread the bacteria onto Muller–Hinton agar. After drying for 10 minutes, antibiotic disks were placed on agar with adequate spacing to prevent the overlap of inhibition zones. The diameters of these zones are measured using a vernier caliper and compared with standardized tables provided by antibiotic disk suppliers to determine minimal inhibitory concentrations (CLSI, 2022) (Fig. 1). ResultsConsistent with culture, microscopy, and the biochemical and VITEK results (Fig. 2), there were (eight) positive counts of Streptococcal isolates obtained from (60) samples from different markets. These (eight) isolates include Streptococcus thoraltensis, Streptococcus sanguinis, Lactococcus lactis subspecies cremoris, and Streptococcus alactolyticus which include (two) isolates of each. In this research, there was a frequency distribution of isolated microorganisms from examined cheese 8 (13.3%) (Table 1). Black, dry crystalline colonies signified strong biofilm formation, while dark colonies without dry crystalline structures suggested intermediate biofilm formation. The pale pink colonies indicate the absence of biofilm formation (Fig. 1). The detailed antibiotic activity results provide a comprehensive overview of the susceptibility patterns of the four Streptococcus species to the tested antibiotics. The susceptibility of S. thoraltensis, S. sanguinis, and S. alactolyticus were (26.6%) and Streptococcus cremoris (20%) to all antibiotics. Resistance of S. thoraltensis was (53%), S. sanguinis, S. alactolyticus were (66%) and S. cremoris was (73%) to all antibiotics (Table 3). Streptococcus thoraltensis, S. cremoris, and S. alactolyticus were susceptible (75%) to Amikacin. S. sanguinis and S. cremoris were susceptible (50%) to Tigecycline, S. thoraltensis, and S. alactolyticus were susceptible (50%) to Doxycycline. Streptococcus sanguinis and S. alactolyticus were susceptible (50%) to Ciprofloxacin. S. thoraltensis and S. sanguinis were susceptible (50%) to Azithromycin. S. sanguinis was susceptible (25%) to vancomycin. Streptococcus alactolyticus was susceptible (25%) to Streptomycin. S. thoraltensis was susceptible (25%) to Amoxicillin and Clavulanic acid. S. cremoris was susceptible (25%) to Penicillin. All isolates were 100% resistant to imipenem, lincomycin, meropenem, methicillin, and chloramphenicol.
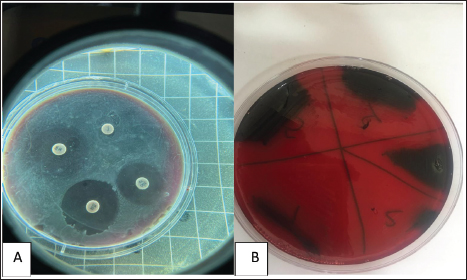
Fig. 1. (A) Antimicrobial susceptibility of Streptococcus isolates on modified Muller-Hinton agar. (B) Streptococcal isolates were cultured in Congo red medium. It is evident that certain antibiotics, such as Tigecycline and Amikacin, show varying degrees of effectiveness against different species, whereas others, like Imipenem and Lincomycin, exhibit consistent resistance across all strains (Tables 2, 4, and 5). DiscussionMozzarella cheese is made specifically from buffalo milk, in addition to other types of animal milk (Araujo et al., 2012). According to Ammin et al. (2024) study, Streptococcus agalactiae and Streptococcus pyogenes were identified as the primary Streptococcus species responsible for pharyngitis in children, with prevalence rates of 24% and 18%, respectively, the research also highlighted the variation in bacterial species distribution based on the patients’ ages. Furthermore, antibiotic sensitivity testing revealed intriguing results; Streptococcus pneumoniae and Streptococcus parasanguinis exhibited high sensitivity to multiple antibiotics, whereas S. pyogenes demonstrated resistance to Metronidazole and Azithromycin (Ammin et al., 2024). Vásquez-García et al. (2017) found isolation from milk buffalo were 11(5.5%). Morea et al. (1999) and Al-Khafaji et al. (2013) examined cheese to investigate the dominant bacteria for the detection of potential pathogens in raw milk-derived unripe cheese. They identified Streptococcus uberis along with other species such as Corynebacterium, Streptococcus, Enterococcus, Staphylococcus, Leuconostoc, and Lactococcus. In Baghdad–Iraq, the first case of vancomycin resistance in raw milk was reported by Al Marjani et al. (2016). The results of sensitivity to vancomycin (25%) were accepted by o Vásquez-García et al. (2017) and noncompatible (12.5%) for penicillin. Bhardwaj et al. (2018) found in India beta-hemolytic Streptococci less resistant to ciprofloxacin (21, 9.5%). On the other hand, all isolates of Enterococcus faecalis in crude milk and imported milk powders at Baghdad markets are resistant to vancomycin (Al-Shammary, 2019). Antimicrobial resistance in Streptococci from animals can differ significantly based on the specific Streptococcal type, so it should not be considered a single entity (Haenni et al., 2018). Antibiotic treatment plays a critical role in managing mastitis infections. Understanding the prevalence of antibiotic resistance and the susceptibility of microorganisms isolated from infected cow milk could improve mastitis treatment strategies. Thus, infection treatment with intramammary antibiotics begins before microbiological culture (Wongkattiya, 2008; Kanaan and AL-Shammary, 2013; Muruzović et al., 2018).
Fig. 2. VITEK results of Streptococcal isolates. Table 1. Distribution of isolated from check cheese (60 samples).
Table 2. Average diameters of antibiotic inhibition zones (mm).
Table 3. Antibiotic susceptibility of isolates.
Table 4. Percentage of antibiotic sensitivity to Streptococcus species.
Table 5. Proportion of antibiotics resistant to streptococcus species.
Antibiotic resistance jeopardizes the effective prevention of infectious diseases. While some bacterial strains are naturally resistant, others develop resistance through mutation, recombination of foreign DNA into their chromosome, or horizontal gene transfer (Brown-Jaque et al., 2015; Petrovich et al., 2020; Zarei-Baygi and Smith, 2021). ConclusionThe primary objective of this study was to conduct a comprehensive microbiological assessment of mozzarella cheese samples, with a specific focus on detecting and evaluating the presence of infective Streptococci. This goal was driven by the need to ensure the highest quality and safety standards in the cheese industry, as well as to address potential health concerns associated with Streptococcal infections. By employing a range of microbiological procedures, we aimed to gain insights into the qualitative aspects of mozzarella cheese, particularly its susceptibility to Streptococcal contamination. The ultimate goal was twofold: first, to establish a robust framework for identifying and characterizing Streptococcal strains in cheese samples, and second, to develop effective strategies for microbiological care and control. The significance of this study lies in its potential to enhance food safety measures and protect public health. Streptococci, particularly certain pathogenic strains, pose serious health risks when present in food products. By understanding the prevalence and behavior of these bacteria in mozzarella cheese, researchers can develop targeted interventions and guidelines to minimize the risk of Streptococcal infections. The findings of this study can contribute to the development of improved cheese manufacturing processes, storage conditions, and sanitation practices. By identifying specific Streptococcal species and their antibiotic susceptibility profiles, the cheese industry can implement tailored measures to prevent contamination and ensure the production of safe and high-quality mozzarella cheese. In conclusion, the ultimate goal of performing microbiological procedures to evaluate mozzarella cheese against infective Streptococci is to safeguard public health, improve food safety standards, and enhance the overall quality of this popular dairy product. The insights gained from this study will undoubtedly benefit both consumers and the cheese industry by fostering a culture of microbiological care and excellence. AcknowledgmentsThe authors thank the zoonoses research unit at the College of Veterinary Medicine, Baghdad University. Conflict of interestThe authors declare no conflict of interest. FundingThe authors declare that finance is personal. Authors’ contributionsThe authors declare that contributions to the research are equal. Data availabilityAll data supporting the research is available within the manuscript. ReferencesAlbanna, A.M.J. and Al-Layla, A.A. 2020. Using real-time PCR to investigate some of antibiotic resistance genes from Streptococcus agalactiae isolates from ewe mastitis cases in Nineveh province. Baghdad Sci. J. 17(3) (Suppl.), 0931. Algboory, H.L., Al-Darwash, A.K., Jarallah, E.M. and Muhialdin, B.J. 2017. Partial purification of antimicrobial peptides from fermented Iraqi camel’s milk by Streptococcus thermophilus and Lactobacillus delbrueckii sp. Bulgaricus. Biochem. Cell. Arch. 17, 697–701. Al-Khafaji, M., Flayyih, M. and Sabah, M. 2013. Isolation, identification and detection of some virulence factors of Staphylococci in milk and cheese in Baghdad. Iraqi J. Sci. 54(4Appendix), 1057–1067. Al Marjani, M.F., Abdul, J., Salman, S., Khudhaier, S.R., Salim, M.Z. and Kadham, Z. A. 2016. First case of vancomycin resistant Streptococcus alactolyticus from raw milk in Baghdad-Iraq. Adv. Environ. Biol. 10, 4–9. Al-Shammary, A.H.A. 2019. Run-off patterns of vancomycin resistant enterococci (VRE clones) in cows’ raw milk and imported milk powders at Baghdad markets. Iraqi J. Vet. Med. 43(2), 61–66. Ammin, R.A., Wadi, H.H., AL-Gburi, E.A.H., Motib, A.S. and Jaber, M.H. 2024. Antibiotic susceptibility of Streptococcus species that cause pharyngitis in children. Diyala J. Med. 26(1), 163–171. Araujo, K.B.S., Rangel, A.H.N., Fonseca, F.C.E., Aguiar, E.M., Simplicio, A.A., Novaes, L.P. and Lima Júnior, D.M. 2012. Influence of the year and calving season on production, composition and mozzarella cheese yield of water buffalo in the State of Rio Grande do Norte, Brazil. Ital. J. Anim. Sci. 11, 87–91. Bhardwaj, N., Mathur, P., Behera, B., Mathur, K., Kapil, A. and Misra, M.C. 2018. Antimicrobial resistance in beta-haemolytic streptococci in India: a four-year study. I.J.M.R. 147(1), 81–87. Blaya, J., Barzideh, Z. and LaPointe, G. 2018. Symposium review: interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment. J. Dairy Sci. 101(4), 3611–3629. Brown-Jaque, M., Calero-Cáceres, W. and Muniesa, M. 2015. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7; doi:10.1016/j.plasmid.2015.01.001 CLSI. 2022. Performance standard for antimicrobial susceptibility testing document M100, 32nd ed. Wayne, PA: Clinical and Laboratory Standards Institute. Coque, T.M., Patterson, J.E., Steckelberg, J.M. and Murray, B.E. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171(5), 1223–1229. De Jong, A., Thomas, V., Klein, U., Marion, H., Moyaert, H., Simjee, S. and Vallé, M. 2013. Pan-European resistance monitoring programmes encompassing food-borne bacteria and target pathogens of food-producing and companion animals. Int. J. Antimicrob. Agents 41(5), 403–409. Devirgiliis, C., Zinno, P. and Perozzi, G. 2013. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front. Microbiol. 4, 301; doi:10.3389/fmicb.2013.00301. Dhanda, V., Chaudhary, P., Toor, D., Kumar, R. and Chakraborti, A. 2013. Antimicrobial susceptibility pattern of beta-haemolytic group A, C and G streptococci isolated from North India. J. Med. Microbiol. 62(3), 386–393. Flayyih, A.S., Hassani, H.H. and Wali, M.H. 2016. Detection of biofilm genes (gtf) in Streptococcus mutans isolated from human dental caries. Iraqi J. Sci. 57(1A), 104–108. Flórez, A.B. and Mayo, B. 2017. Antibiotic resistance-susceptibility profiles of Streptococcus thermophilus isolated from raw milk and genome analysis of the genetic basis of acquired resistances. Front. Microbiol. 8, 315724. Foster, G., Kirchner, M., Muchowski, J., Duggett, N., Randall, L., Knight, H.I. and Whatmore, A.M. 2020. Streptococcus caledonicus sp. nov., isolated from sheep. Int. J. Syst. Evol. Microbiol. 70, 2611–2615; doi:10.1099/ijsem.0.004081. Freeman, D.J., Falkiner, F.R. and Keane, C.T. 1989. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42(8), 872–874. Gad, G.F.M., Abdel-Hamid, A.M. and Farag, Z.S.H. 2014. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz. J. Microbiol. 45, 25–33. Garnier, L., Valence, F. and Mounier, J. 2017. Diversity and control of spoilage fungi in dairy products: an update. Microorganisms 5(3), 42. Haenni, M., Lupo, A. and Madec, J.Y. 2018. Antimicrobial resistance in Streptococcus spp. Microbiol. Spectr. 6(2), 10–1128. Hu, Y., Yang, X., Qin, J., Lu, N., Cheng, G., Wu, N. and Zhu, B. 2013. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4(1), 2151. Hussaini, S.Z., Shaker, M., Gulve, R.M. and Iqbal, M.A. 2014. Bacterial analysis of raw and packed milk of Beed City. J.A.S.A.T. 1(1), 53–58. Hussein, R.M. and Jabbar, A.K.N. 2020. Isolation and identification of zoonotic importance bacteria from milk, milk products and human in Diyala, Iraq. Plant Arch. 20(2), 6809–6817. Kanaan, M.H.G. and AL-Shammary, A.H.A. 2013. Detection of methicillin or multidrug resistant Staphylococcus aureus (MRSA) in locally produced raw milk and soft cheese in Baghdad markets. Iraqi J. Vet. Med. 37(2), 226–231. Ledenbach, L.H. and Marshall, R.T. 2009. Microbiological spoilage of dairy products. In Compendium of the microbiological spoilage of foods and beverages. Eds., Sperber, W. and Doyle, M. New York, NY: Springer, pp: 41–67. Losito, F., Arienzo, A., Bottini, G., Priolisi, F.R., Mari, A. and Antonini, G. 2014. Microbiological safety and quality of Mozzarella cheese assessed by the microbiological survey method. J. Dairy Sci. 97(1), 46–55. Lu, B., Fang, Y., Huang, L., Diao, B., Du, X., Kan, B. and Wang, D. 2016. Molecular characterization and antibiotic resistance of clinical Streptococcus dysgalactiae subsp. equisimilis in Beijing, China. Infect. Genet. Evol. 40, 119–125. Marshall, B.M. and Levy, S.B. 2011. Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733; doi:10.1128/CMR. 00002-11. Mazzoli, R., Bosco, F., Mizrahi, I., Bayer, E.A. and Pessione, E. 2014. Towards lactic acid bacteria-based biorefineries. Biotechnol. Adv. 32(7), 1216–1236. Morea, M., Baruzzi, F. and Cocconcelli, P.S. 1999. Molecular and physiological characterization of dominant bacterial populations in traditional Mozzarella cheese processing. J. Appl. Microbiol. 87(4), 574–582. Mounam, M.A.A., Al-Gburi, N.M. and Razook, B.R. 2023. Investigation of pathogenic Streptococci species in local and imported mozzarella soft cheese in Baghdad city. Adv. Life Sci. 10(3), 486–490. Muruzović, M.Ž., Mladenović, K.G., Žugić-Petrović, T.D. and Čomić, L.R. 2018. In vitro evaluation of the antimicrobial potential of Streptococcus uberis isolated from a local cheese from Southeastern Serbia. Vet. Arh. 88(4), 521–534. Motaweq, Z.Y. and Naher, H.S. 2017. Antimicrobial susceptibility of Streptococcus pneumoniae isolates causing LRTI in Najaf, Iraq. Environ. Socio-Econom. Stud. 5(2), 10–18. Najim, N.H. 2010. Influence of mild pulsed electric field conditions on the growth and protease activity of Streptococcus thermophilus. Iraqi J. Vet. Med. 34(2), 20–29. Pal, M., Mulu, S., Tekle, M., Pintoo, S.V. and Prajapati, J. 2016. Bacterial contamination of dairy products. Beverage Food World 43(9), 40–43. Petrovich, M.L., Zilberman, A., Kaplan, A., Eliraz, G R., Wang, Y., Langenfeld, K. and Wells, G.F. 2020. Microbial and viral communities and their antibiotic resistance genes throughout a hospital wastewater treatment system. Front. Microbiol. 11, 153. Rauh, V. and Xiao, Y. 2022. The shelf life of heat-treated dairy products. Int. Dairy J. 125, 105235. Ryu, S.H., Lee, J.H., Park, S.H., Song, M.O., Park, S.H., Jung, H.W. and Lee, Y.K. 2012. Antimicrobial resistance profiles among Escherichia coli strains isolated from commercial and cooked foods. Int. J. food Microbial. 159(3), 263–266. Soares-Santos, V., Barreto, A.S. and Semedo-Lemsaddek, T. 2015. Characterization of enterococci from food and food-related settings. J. Food Prot. 78, 1320–1326; doi:10.4315/0362-028X.JFP-14-419. Taher, E.M., Hemmatzadeh, F., Aly, S.A., Elesswy, H.A. and Petrovski, K.R. 2020. Molecular characterization of antimicrobial resistance genes on farms and in commercial milk with emphasis on the effect of currently practiced heat treatments on viable but nonculturable formation. J. Dairy Sci. 103(11), 9936–9945. Tian, Z., Lu, S., Jin, D., Yang, J., Pu, J., Lai, X.H. and Xu, J. 2019. Streptococcus chenjunshii sp. nov. isolated from feces of Tibetan antelopes. Int. J. Syst. Evol. Microbiol. 69(4), 1237–1243. Tsakalidou, E., Zoidou, E., Pot, B., Wassill, L., Ludwig, W., Devriese, L.A. and Kersters, K. 1998. Identification of streptococci from Greek Kasseri cheese and description of Streptococcus macedonicus sp. nov. Int. J. Syst. Evol. Microbiol. 48(2), 519–527. Vásquez-García, A., Silva, T.D.S., Almeida-Queiroz, S.R.D., Godoy, S.H., Fernandes, A.M., Sousa, R.L. and Franzolin, R. 2017. Species identification and antimicrobial susceptibility profile of bacteria causing subclinical mastitis in buffalo. Pesqui. Vet. Bras. 37, 447–452. Verraes, C., Claeys, W., Cardoen, S., Daube, G., De Zutter, L., Imberechts, H. and Herman, L. 2014. A review of the microbiological hazards of raw milk from animal species other than cows. Int. Dairy J. 39(1), 121–130. Wang, H., McEntire, J.C., Zhang, L., Li, X. and Doyle, M. 2012. The transfer of antibiotic resistance from food to humans: facts, implications and future directions. Rev. Sci. Technol. 31, 249–260; doi:10.20506/rst.31 1.2117. Watkins, R.R. and Bonomo, R.A. 2016. Overview: global and local impact of antibiotic resistance. Infect. Dis. Clin. 30(2), 313–322. Wongkattiya, N. 2008. Characterization and biological control of bovine of Streptococcus uberis strains associated with bovine mastitis. PhD Thesis, School of Applied Sciences, RMIT University, Melbourne, Australia. Yao, J., Gao, J., Guo, J., Wang, H., Zhang, E.N., Lin, Y. and Tao, S. 2022. Characterization of bacteria and antibiotic resistance in commercially produced cheeses sold in China. J. Food Prot. 85(3), 484–493. Zarei-Baygi, A. and Smith, A.L. 2021. Intracellular versus extracellular antibiotic resistance genes in the environment: prevalence, horizontal transfer, and mitigation strategies. Bioresour. Technol. 319, 124181. Zhang, Y.J., Hu, H.W., Chen, Q.L., Singh, B.K., Yan, H., Chen, D. and He, J.Z. 2019. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 130, 104912. | ||
| How to Cite this Article |
| Pubmed Style Mounam MAA, Razook BR, Madi EH. Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad. Open Vet. J.. 2025; 15(3): 1178-1186. doi:10.5455/OVJ.2025.v15.i3.10 Web Style Mounam MAA, Razook BR, Madi EH. Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad. https://www.openveterinaryjournal.com/?mno=228563 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i3.10 AMA (American Medical Association) Style Mounam MAA, Razook BR, Madi EH. Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad. Open Vet. J.. 2025; 15(3): 1178-1186. doi:10.5455/OVJ.2025.v15.i3.10 Vancouver/ICMJE Style Mounam MAA, Razook BR, Madi EH. Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad. Open Vet. J.. (2025), [cited January 25, 2026]; 15(3): 1178-1186. doi:10.5455/OVJ.2025.v15.i3.10 Harvard Style Mounam, M. A. A., Razook, . B. R. & Madi, . E. H. (2025) Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad. Open Vet. J., 15 (3), 1178-1186. doi:10.5455/OVJ.2025.v15.i3.10 Turabian Style Mounam, Moutaz A.w. Abdul, Basil R.f. Razook, and Entesar Hussain Madi. 2025. Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad. Open Veterinary Journal, 15 (3), 1178-1186. doi:10.5455/OVJ.2025.v15.i3.10 Chicago Style Mounam, Moutaz A.w. Abdul, Basil R.f. Razook, and Entesar Hussain Madi. "Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad." Open Veterinary Journal 15 (2025), 1178-1186. doi:10.5455/OVJ.2025.v15.i3.10 MLA (The Modern Language Association) Style Mounam, Moutaz A.w. Abdul, Basil R.f. Razook, and Entesar Hussain Madi. "Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad." Open Veterinary Journal 15.3 (2025), 1178-1186. Print. doi:10.5455/OVJ.2025.v15.i3.10 APA (American Psychological Association) Style Mounam, M. A. A., Razook, . B. R. & Madi, . E. H. (2025) Antimicrobial susceptibility patterns of infective Streptococci recovered from local and imported mozzarella cheese from different markets in Baghdad. Open Veterinary Journal, 15 (3), 1178-1186. doi:10.5455/OVJ.2025.v15.i3.10 |