| Short Communication | ||
Open Vet. J.. 2025; 15(1): 456-464 Open Veterinary Journal, (2025), Vol. 15(1): 456-464 Short communication Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication studyLily Arsanti Lestari1,2, Wan Nurul Aini1,3, Marlyn Dian Laksitorini1,3, Yuny Erwanto1,4, Agustina Ari Murti Budi Hastuti1,3, Mohammad Zainal Abidin1,4, Nurulia Hidayah5, Muhammad Jefriyanto Budikafa6 and Abdul Rohman1,3*1Center of Excellence, Institute for Halal Industry and System (IHIS), Universitas Gadjah Mada, Yogyakarta, Indonesia 2Department of Nutrition and Health, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, Indonesia 3Faculty of Pharmacy, Universitas Gadjah Mada, Yogyakarta, Indonesia 4Faculty of Animal Science, Universitas Gadjah Mada, Yogyakarta, Indonesia 5Field Research Center, Universitas Gadjah Mada, Yogyakarta, Indonesia 6Faculty of Islamic Economics and Business, State Islamic University Prof. K. H. Saifuddin Zuhri, Purwokerto, Indonesia *Corresponding Author: Abdul Rohman. Center of Excellence, Institute for Halal Industry and System (IHIS), Universitas Gadjah Mada, Yogyakarta, Indonesia. Email: abdul_kimfar [at] ugm.ac.id Submitted: 16/09/2024 Accepted: 26/12/2024 Published: 31/01/2025 © 2025 Open Veterinary Journal
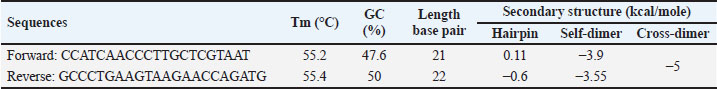
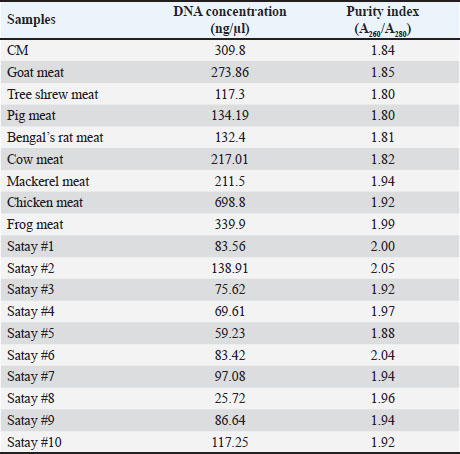
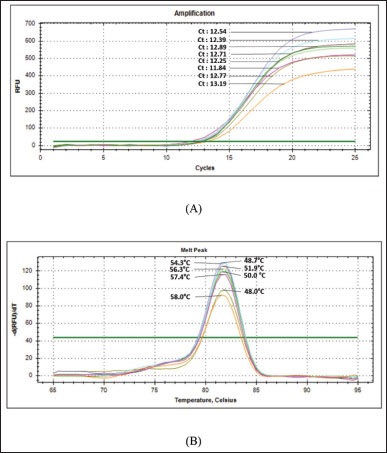
AbstractBackground: Canine meat (CM) is one of the non-halal meats prohibited for consumption by the Muslim community. Due to its low prices compared with beef, CM is typically used as meat adulterants in halal food-based products such as Satay and meatballs to get economic profits. Aim: The objective of this study was to design a novel species-specific primer in combination with real-time polymerase chain reaction for analysis of Canine’s DNA for halal authentication analysis. Methods: A Primer targeting the D-loop region of mitochondrial DNA was designed and subjected to a validation procedure by assessing some performance characteristics including specificity, amplification efficiency (E), sensitivity, repeatability, and linearity describing the correlation between the concentration of Canine’s DNA (x-axis) and quantification cycle (Cq) in y-axis. The designed primer was specific over other meat DNAs applying the annealing temperature (Tm) of 57.8°C. Results: The Real-Time Polymerase Chain Reaction (RT-PCR) method produced an acceptable amplification efficiency (E) of 109.7% with the coefficient of determination (R2) for the correlation between Cq and log DNA concentration of 0.999. The sensitivity of the developed method provides a limit of detection (LoD) value of 31.25 pg/µl. The precision of the analytical method is acceptable with a relative standard deviation value of 2%. The method with the designed D-loop primer was successfully applied for the detection and quantification of Canine’s DNA in food products. There are no amplification profiles for Canine DNA in marketed goat’s satay products. Conclusion: RT-PCR combined with a novel primer targeting D-loop provides a specific and accurate analytical tool for the identification of CM for halal authentication studies. Keywords: D-loop primer, Real time-PCR, Canine meat, Goat’s satay. IntroductionIn recent years, the substitution of halal meats having high prices in the market such as beef with non-halal meats with lower prices such as pork, rat meat, and dog meat has been reported in some scientific literature (Pebriana et al., 2017). Erwanto et al. (2014) reported that pork contamination was found at 9 of 20 meatball stalls in Yogyakarta regions. This action is motivated by economic reasons in which halal meats are more expensive than non-halal meats, or even non-halal meats can be freely available such as canine meats (CMs) (Kurniasih et al., 2019). According to Indonesia’s National Strategic Food Price Information Center, the price of beef is 130 k/kg, while the price of CMs at the slaughterhouse starts at 30 k–50 k/kg. One of the favorite meat-based food products in Indonesia is Satay, and Satay’s name is typically derived from meat sources such as Beef’s Satay, Chicken’s Satay, and Goat’s satay. As a popular grilled food delicacy, Satay is also popular in Southeast Asian regions including Malaysia, Thailand, and Singapore. Satay is similar to shish kebab which is popular in Turkey and in Middle Eastern countries (Jinap et al., 2013). Among these Satay, goat’s Satay is the most consumed by Indonesian people (Irnanda et al., 2012), as a consequence, goat’s meat is the potential to be adulterated or substituted with non-halal meats. For this reason, the availability of analytical methods capable of confirming the presence of non-halal meats is very urgent. Identification of meat types in meat-based food products is very urgent, especially for the identification of non-halal meats. Indonesia has implemented the Indonesian Act No. 33 the year 2014 on Halal Products Assurance stating that all products including meat-based products distributed and sold in Indonesia must be halal-certified, and the products suspected to contain non-halal meats must be subjected to testing in an accredited laboratory according to ISO/IEC 17025: 2017 (Malik et al., 2016). Several analytical methods have been proposed and used for the identification, detection, and confirmation of non-halal meats in food products, mainly based on molecular spectroscopic methods including Fourier Transform Infrared (FTIR) spectroscopic (Kuswandi et al., 2015), Nuclear Magnetic Resonance (NMR) spectroscopy (Fadzillah et al., 2017) and Raman spectroscopy in combination with multivariate data analysis (Mortas et al., 2022) and chromatographic-based methods including gas chromatography hyphenated with mass spectrometer or GC-MS (Indrasti et al., 2010) and liquid chromatography hyphenated with mass spectrometer or LC-MS/MS (Abbas et al., 2020). In addition, some screening methods for rapid detection of non-halal meats are also proposed such as electronic nose (Nurjuliana et al., 2011), electronic tongue (Tian et al., 2019), and differential scanning calorimetry or DSC (Bertram et al., 2006). All these methods need advanced data treatment which involves big data analysis, therefore some methods based on specific markers such as protein and DNA-based methods are widely used. DNA-based methods using polymerase chain reaction are currently taken into account as a method of choice or gold methods for identification and confirmation of non-halal meats in food products due to their specificity and sensitivity. Numerous PCR methods have been reported for the identification of non-halal meats in food products including real-time PCR and conventional PCR using species-specific primers (SSPs), singleplex or multiplex PCR, random amplified polymorphic DNA, and restriction fragment length polymorphisms as reviewed by some authors (Salihah et al., 2016; Rohman et al., 2020a,b; Muflihah et al ., 2023). Real-time PCR and conventional PCR using SSP targeting on mitochondrial, cyt-b, and d-loop and other PCR techniques have been successfully applied for analysis of non-halal meat-based products such as analysis of pork in meatball (Erwanto et al., 2012), analysis of pork in abon (Rahmawati et al., 2016), analysis of dog meat in meatball (Ali et al., 2015), and analysis of rat meat in meatball (Cahyadi et al., 2020). D-loop has good specificity in identifying raw materials in food products which has been processed through cooking and degradation (Kusnadi et al., 2020). Many studies related to food identification using D-loop primers have been conducted, such as analysis of pork in sausages (Maulani et al ., 2020), analysis of pork gelatin in capsule shells (Sudjadi et al., 2016), and analysis of monkey meat contamination in beef meatballs (Kurniawati et al., 2024). The use of D-loops in detecting foods is considered to have a high success rate, so it has a high opportunity to be used in halal detection methods with various PCR-based methods (Sosiawan et al., 2020). From literature searching, the identification of non-halal meats in Satay is very limited. Therefore, the objective of this study was to design SSP followed by real-time PCR analysis for the identification of CM in Goat’s Satay. In addition, the developed real-time PCR using SSP targeting on D-loop mitochondria was validated by determining some performance characteristics which included primer specificity, sensitivity, extraction efficiency, reproducibility, and application of real-time PCR for analysis of Satay commercially available in the market. Materials and MethodsExperimental details and treatmentCM and rat meat were obtained from markets and farmers. The other types of meats namely goat meat, mackerel, pork, chicken, beef, and frog meat were purchased from traditional markets in Yogyakarta Province, Indonesia. Ten commercial goat’s satay samples were collected from Yogyakarta, Indonesia. Some sets of primers for real-time PCR analysis were purchased from Genetika Science (Indonesia). Primer designingA set of primers targeting D-loop was designed using software from Integrated DNA Technologies (IDT, California, USA). DNA sequence of Canis lupus familiaris mitochondrion DNA complete genome (MW549038.1) was obtained from NCBI (http://www.ncbi.nlm.nih.gov). A set of primers was selected from five primers candidates from IDT after in silico specificity test using BLASTn (Basic Local Alignment Search Tool Nucleotide) from the NCBI website by comparing canine species with goat, rat, mackerel, pig, frog, cow, chicken, cow, and tree shrew. The selection of primer also considers the presence of the predicted secondary structure of oligonucleotides using OligoAnalyser Tool from IDT Software. The selected primer for further analysis and validation is shown in Table 1. DNA isolationThe isolation of DNAs from raw meat and Satay products were isolated using FavorPrepTM Tissue Genomic DNA Extraction Mini Kit (Favorgen, Taiwan). The procedure of isolation followed the manufacture’s instruction with several stages, including cell lysis using buffer lysis and proteinase-K, binding DNA with a silica column matrix, washing DNA from a contaminant, and eluting DNA from the silica column. Analysis using real-time PCRReal-time PCR amplification was carried out using PCR CFX96 (Biorad, USA) with a total volume of 10 µl containing 5 µl of 2x SensiFAST SYBR® No-Rox Mix (Meridian Bioscience, USA), 0.4 µl of each forward and reverse primers (10 µM), 1 µl template DNA with concentration 25 ng/ µl and 3.2 µl nuclease-free water. The reaction was carried out in 25 amplification cycles on the program with cycling conditions used was as follows: pre-denaturation at 95ºC for 3 minutes, denaturation at 95ºC for 5 seconds, annealing step at 57.8ºC for 10 seconds, and elongation step at 72ºC for 20 seconds. Validation of real-time PCR methodsReal-time PCR using an SSP primer previously designed was subjected to validation by determining several parameters, including specificity, efficiency, sensitivity, and repeatability as in Codex Alimentarius Commission (CAC) (Aina et al., 2019). The validated methods were then employed for the identification of canine DNA in commercial goat’s Satays by amplifying real-time PCR methods using primer D-loop 38 for analysis of 10 commercial goat’s satay samples with canine’s DNA as positive control and no template control (NTC) as negative control. Data analysisThe isolated DNAs were subjected to the purity assessment and the concentration of DNAs was quantified using Nano Quant Spark Tecan (Switzerland) at Wavelengths of 260 and 280 nm. Selection of the optimum annealing temperature was carried out by looking at the increase of the curve and the cycle threshold (Ct) value of the amplification curve. The specificity test of primer D-loop was evaluated by amplifying DNAs isolated from several types of meats (canine, goat, cow, pig, mackerel, Bengal rat, tree shrew, chicken, and frog) and NTC. The sensitivity of real-time PCR using D-loop was carried out by determining the limit of detection (LOD), as the lowest quantity or concentration of DNA that still can be reliably detected and amplified with reproducible Ct using real-time PCR. The LOD value was evaluated using several dilutions of the isolated DNAs from raw CM and then amplified with a designed primer. The amplification efficiency (E-value), slopes, and coefficient of determination (R2) were calculated using the software Bio-Rad CFX manager. Efficiently was aimed to know the capability of the real-time PCR method in amplifying the DNA targets and it was determined from standard curve plotting Ct values (y-axis) versus log concentration of DNA (x-axis). The repeatability of the validated method was carried out using eight replicates. Results and DiscussionsIn this study, the SSP was specifically designed using NCBI software for the identification of CM in processed foods, especially in Satay products using real-time PCR methods. The primer was designed to targeting D-loop fragment of mitochondrial DNA (mtDNA) C. familiaris. Some advantages of using D-loop fragment are that it has the highest levels of polymorphism in mtDNA and has good specificity in the identification of food processing products (Liu et al., 2019), while the selection of mtDNA is due to the presence of mtDNA in hundreds to thousands of copies per cell and it will be advantageous in analysis of very limited number of samples (Weedn et al., 2018). The DNA templates were successfully isolated from raw meats and Satay products using FavorPrepTM Tissue Genomic DNA Extraction Mini Kit. Analysis of DNA’s purity and concentration was determined using absorbance ratios at wavelength 260 and 280 (A260/280) using the instrument of Tecan’s NanoQuant Plate. The ratio values at 260 and 280 (A260/280) can be used as a purity index of DNA in which DNAs with A260/280 of 1.8–2.0 are considered pure and it is suitable to be used as a DNA template during real-time PCR analysis. In addition, A260/280 <1.8 indicates that the isolated DNAs also contain protein, phenols, and carbohydrates, while A260/280 > 2.0 indicates the isolated DNA contains RNA (Orbayinah et al., 2020). Table 2 shows the purity indexes and concentrations of DNA isolated from raw meat and commercial Satay products. The isolated DNAs had purity indexes approaching 1.8–1.99 indicating that the isolated DNAs are pure and suitable to be used as DNA templates for further analysis using real-time PCR analysis. The optimization of annealing temperatures (Ta) is an important factor to be considered during analysis using real-time PCR, since Ta is associated with the attachment of primer in DNA targets. The gradient temperature was evaluated to find the optimal Ta of the designed primer. A range of Ta of 48ºC–58ºC was examined, and finally, Ta of 57.4ºC was selected as the optimal temperature due to its capability to provide the lowest values of Ct as shown in Figure 1A. From melting curve analysis (MCA), the suitability of 57.4ºC as annealing temperature was supported by the presence of a single peak in the amplification curve which means that there were no prime dimers and non-specific products observed during PCR amplification (Fig. 1B). Furthermore, this Ta was used for further analysis including validation of real-time PCR and the use of validated methods for analysis of commercial Satay products. Table 1. Primer D-loop 38 targeting mitochondrial D-loop fragment of C. familiaris.
Table 2. The concentration and purity of DNA isolated from raw meat and commercial goat’s satay sample.
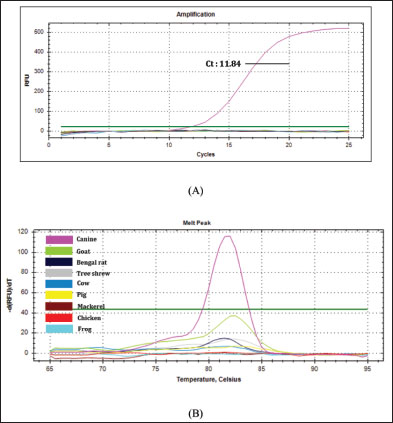
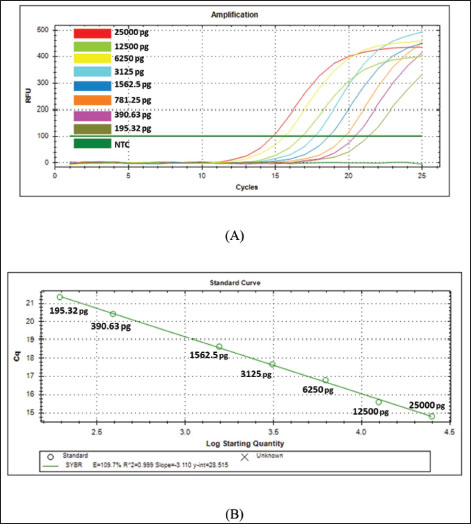
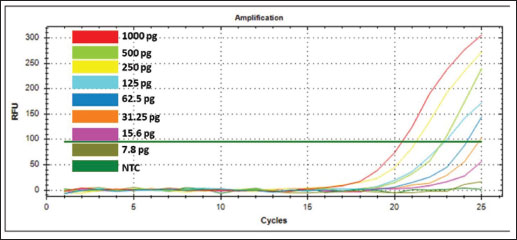
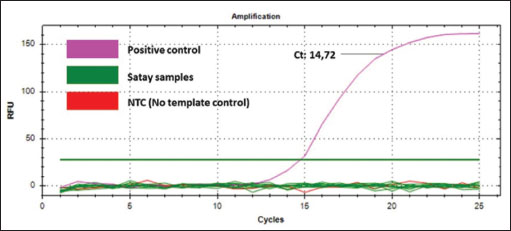
The validation method of real-time PCR method using SSP targeting on D-loop 38 was carried out by assessing some validation parameters which include primer specificity, efficiency, sensitivity, and repeatability. Primer specificity test was carried out using optimal Ta (57.4ºC) by amplifying the isolated DNAs from different animal tissues (canine, goat, cow, Bengal’s rat, chicken, frog, mackerel, pork, and tree shrew) along with negative control or NTC. Figure 2A exhibited the amplification curve of real-time PCR using primer D-loop, in which only DNAs isolated from CM could be amplified with Ct value of 11.84 and relative fluorescence unit of 521. Figure 2B exhibited the MCA of the amplification curve in which only a single peak was observed above the threshold line belonging to CM. From the amplification curve and MCA, it can be concluded that SSP targeting on D-loop was specific for DNAs isolated from CM and the primer did not amplify other DNAs used. Efficiency and sensitivity tests were carried out by preparing a 2-fold serial dilution of pure canine DNA and followed by making a standard curve correlating between log concentration of DNA (x-axis) and Ct-values (y-axis) to find the efficiency (E-value). The amplification profile of DNAs isolated from CM with 8 different concentrations is shown in Figure 3A. A good linear relationship existed with R2 of 0.999, the slope of −3.110, y-intercept of 28.515, with efficiency value (E) 109.7% (Fig. 3B). CAC (CAS, 2010) declared the acceptance criteria for validation parameters of quantitative PCR assay is R2 ≥0.98 and E value of 90%–110% (Rohman et al., 2020). Therefore, the E-value and R2 met the requirements. LOD value was analyzed from eight different concentrations of DNA (25,000; 12,500; 6,250; 3,125; 1,562.5; 781.25; 390.63; 195.32 pg/µl), and then the amplification curve was obtained. The smallest concentration (195.32 pg/ µl) can be still amplified by Real-time PCR above the threshold limit, therefore, DNAs with concentrations of 1,000, 500, 250, 125, 62.5, 31.25, 15.6, and 7.8 pg/ µl were used. From the amplification curve, the LOD value obtained is 31.25 pg/µl, as shown in Figure 4. The precision of real time PCR was evaluated by repeatability (intra-assay) test. The DNAs isolated from CM at a concentration of 6,250 pg/µl were amplified using SSP D-loop primer 38, and the Ct values were evaluated from eight replicates. The relative standard deviation (RSD) was used for the evaluation of repeatability, and RSD values of Ct were found to be 2% which is lower than the maximum RSD values set by CAC, i.e., ≤25%, therefore the RSD values met the acceptance criteria for precision parameter. The validated real-time PCR method using primer D-loop 38 was then applied for the detection of CM in 10 commercial goat’s Satay around Yogyakarta, Indonesia, as shown in Figure 5. There were 10 samples along with positive control (CM’s DNA) and NTC evaluated, and this validated method shows that only the DNA template of CM (positive control) was amplified with Ct value of 14.72, while all the commercial goat’s satay and NTC (negative control) shows no amplification response. It can be concluded that all the evaluated commercial goat’s satay samples did not contain CM.
Fig. 1. Amplification curve during the optimization of annealing temperature at different temperatures. (A)=Amplification curve; (B)=melt peak curve.
Fig. 2. The specificity test of primer D-loop 38 using DNAs of CM and 8 other meat species. (A)=Amplification curves; (B)=Melting peak curve.
Fig. 3. Real-time PCR amplification using primer D-loop 38 at different DNA concentrations of CM with serial dilutions. (A)=Amplification curves; (B)=Standard curve correlating between log DNA concentration (x=axis) and Ct (y-axis).
Fig. 4. Amplification of Canine’s DNA at different DNA concentrations with serial dilution for sensitivity test using primer D-loop 38 of C. familiaris.
Fig. 5. The amplification curve from real-time PCR methods using primer D-loop 38 for analysis of 10 commercial goat’s satay samples. The Realtime PCR methods using primer D-loop 38 have been successfully developed and validated to identify and confirm the presence of CM in commercial goat’s satay. The design used in this method allows it to be further developed to detect other non-halal meat such as rats, pigs, monkeys, and so on. In addition, this method can also be developed in combination with other methods or approaches such as metabolomic approaches to improve the ability to detect products which has more complex characteristics like pharmaceutical products and cosmetic products. ConclusionThe SSP targeting mitochondrial D-Loop 38 could specifically amplify DNAs isolated from CM using the optimized annealing temperature of 57.4ºC. Real-time using this primer was valid as indicated by accepted validation criteria. This method is sensitive enabling the detection of canine’s DNAs as low as 31.25 pg/µl DNA. This validated method shows canine’s DNA was not found in all commercial goat’s satay samples. The development RT-PCR methods using primer D-loop 38 can be used for the identification and confirmation of CM in food products to support the development of a standard method for halal authentication analysis. AcknowledgmentThe authors thank Direktorat Penelitian UGM for the financial support through the research grant of Learning Center in Collaboration (LC-IC) UGM with contract number: 4799/UN1/DITLIT/PT.01.03/2024 awarded to Dr. Lily Arsanti Lestari. Conflict of interestThe authors declare that there is no conflict of interest. FundingThis research was supported by Direktorat Penelitian UGM through the research grant of Learning Center in Collaboration (LC-IC) UGM with contract number: 4799/UN1/DITLIT/PT.01.03/2024 awarded to Dr. Lily Arsanti Lestari. Authors’ contributionsLAL: Conceptualized the research, designed the experiments, reviewing the manuscript; WNA: Collected the data, performed the experiments, conducted statistical analysis, wrote the manuscript and participated in manuscript revisions. MDL: Conceptualized the research, designed the experiments, wrote the manuscript and participated in manuscript revisions; YE: Conceptualized the research, designed the experiments, wrote the manuscript and participated in manuscript revisions; AAMBH: Designed the experiments, wrote the manuscript and participated in manuscript revisions; MZA: Designed the experiments, wrote the manuscript and participated in manuscript revisions; NH: Designed the experiments, wrote the manuscript and participated in manuscript revisions; MJB: edited the paper and participated in manuscript revisions. AR: Conceptualized the research, designed experiments, supervised students, edited the paper and participated in manuscript revisions. Data availabilityAll data supporting the findings of this study are available within the manuscript. ReferencesAbbas, N., Ali, A., Kumari, S., Iqbal, A., Husain, A., Saeed, T., AbdulAmer Al-Ballam, Z., Ahmed, N., El-Seedi, H.R. and Musharraf, S.G. 2020. Untargeted-metabolomics differentiation between poultry samples slaughtered with and without detaching spinal cord. Arab. J. Chem. 13(2), 9081–9089. Aina, G.Q., Erwanto, Y., Hossain, M., Johan, M.R., Ali, M.E. and Rohman, A. 2019. The employment of q-PCR using specific primer targeting on mitochondrial cytochrome-b gene for identification of wild boar meat in meatball samples. J. Adv. Vet. Anim. Res. 6(3), 300–307. Ali, M.E., Razzak, M.A., Hamid, S.B.A., Rahman, M.M., Al Amin, M., Rashid, N.R.A. and Asing, 2015. Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods. Food. Chem. 177, 214–224. Bertram, H.C., Wu, Z., van den Berg, F. and Andersen, H.J. 2006. NMR relaxometry and differential scanning calorimetry during meat cooking. Meat Sci. 74(4), 684–689. Cahyadi, M., Wibowo, T., Pramono, A. and Abdurrahman, Z.H., 2020. A novel multiplex-pcr assay to detect three non-halal meats contained in meatball using mitochondrial 12s rrna gene. Food. Sci. Anim. Resour. 40(4), 628–635. Erwanto, Y., Abidin, M.Z., Muslim, E.Y.P., Sugiyono, S. and Rohman, A. 2014. Identification of pork contamination in meatballs of Indonesia local market using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis. Asian-Aust. J. Anim. Sci. 27(10), 1487–1492. Erwanto, Y., Abidin, M.Z., Sismindari, X. and Rohman, A., 2012. Pig species identification in meatballs using polymerase chain reaction- restriction fragment length polymorphism for halal authentication. Int. Food Res. J. 19(3), 901–906. Fadzillah, N.A., Rohman, A., Salleh, R.A., Amin, I., Shuhaimi, M., Farahwahida, M.Y., Rashidi, O., Aizat, J.M. and Khatib, A., 2017. Authentication of butter from lard adulteration using high-resolution of nuclear magnetic resonance spectroscopy and high-performance liquid chromatography. Int. J. Food. Prop. 20(9), 2147–2156. Indrasti, D., Che Man, Y.B., Mustafa, S. and Hashim, D.M. 2010. Lard detection based on fatty acids profile using comprehensive gas chromatography hyphenated with time-of-flight mass spectrometry. Food Chem. 122(4), 1273–1277. Irnanda, K., Meiftasari, A., Nagadi, S. and Lukitaningsih, E., 2012. Safety evaluation of chicken satay in Yogyakarta Indonesia based on Benzo[a]pyrene content. Ind. J. Cancer Chemoprev. 3(3), 432. Jinap, S., Mohd-Mokhtar, M.S., Farhadian, A., Hasnol, N.D.S., Jaafar, S.N. and Hajeb, P. 2013. Effects of varying degrees of doneness on the formation of heterocyclic aromatic amines in chicken and beef satay. Meat Sci. 94(2), 202–207. Kurniasih, K.S.I., Hikmah, N., Erwanto, Y. and Rohman, A. 2019. Qualitative and quantitative analysis of canine ( Canis lupus familiaris ) meat in meatballs for halal authentication study using real-time polymerase chain reaction. Int. J. Agric. Biol. 23(1), 103–108 Kurniawati, D.R., Sismindari, S., Rumiyati, R., Wibowo, F.S., Pebriyanti, N.W., Irnawati, I. and Rohman, A. 2024. Specific real-time PCR assay targeting d-loop gene and short amplicon sequencing for identification of monkey meat in beef meatballs. Ind. J. Chem. 24(2), 315–324. Kusnadi, J., Ashari, N.A. and Arumingtyas, E.L. 2020. Specificity of various mitochondrial DNA (mtDNA), ND5, D-Loop, and Cty-b DNA primers in detecting pig (Sus scrofa) DNA fragments. Am. J. Mol. Biol. 10, 141–147. Kuswandi, B., Putri, F.K., Gani, A.A. and Ahmad, M. 2015. Application of class-modelling techniques to infrared spectra for analysis of pork adulteration in beef jerkys. J. Food Sci. Technol. 52(12), 7655–7668. Liu, W., Tao, J., Xue, M., Ji, J., Zhang, Y., Zhang, L. and Sun, W. 2019. A multiplex PCR method mediated by universal primers for the identification of eight meat ingredients in food products. Eur. Food Res. Tech. 245, 2385–2392. Malik, A., Sutantyo, M.L., Hapsari, I., Sinurat, A.V., Purwati, E.M., Jufri, M. and Suryadi, H. 2016. Simultaneous identification and verification of gelatin type in capsule shells by electrophoresis and polymerase chain reaction. J. Pharm. Investig. 46(5), 475–485. Maulani, T.R., Susilo, H., Indriati, M. and Suhaemi, A. 2020. Deteksi Cemaran DNA Babi Dengan RT-PCR Pada Sosis Tanpa Logo Halal MUI Dari Empat Kecamatan di Kabupaten Pandeglang Banten. Gorontalo Agric. Technol. J. 3(2), 72. Mortas, M., Awad, N. and Ayvaz, H. 2022. Adulteration detection technologies used for halal/kosher food products: an overview. Discover Food. 2, 15. Muflihah, Hardianto, A., Kusumaningtyas, P., Prabowo, S. and Hartati, Y.W. 2023. DNA-based detection of pork content in food. Heliyon 9(3), e14418. Nurjuliana, M., Che Man, Y.B., Mat Hashim, D. and Mohamed, A.K.S. 2011. Rapid identification of pork for halal authentication using the electronic nose and gas chromatography mass spectrometer with headspace analyzer. Meat Sci. 88(4), 638–644. Orbayinah, S., Hermawan, A. and Sismindari, Rohman, A. 2020. Detection of pork in meatballs using probe taqman real-time polymerase chain reaction. Food Res. 4(5), 1563–1568. Pebriana, R.B., Rohman, A., Lukitaningsih, E. and Sudjadi, 2017. Development of FTIR spectroscopy in combination with chemometrics for analysis of rat meat in beef sausage employing three lipid extraction systems. Int. J. Food Prop. 20(S2), 1995–2005. Rahmawati, Sismindari, Raharjo, T.J., Sudjadi. and Rohman, A. 2016. Analysis of pork contamination in Abon using mitochondrial D-Loop22 primers using real time polymerase chain reaction method. Int. Food Res. J. 23(1), 370–374. Rohman, A., Pebriyanti, N.W., Sismindari, Windarsih, A., Ramadhani, D., Larasati, R. and Yulisa, H. 2020a. Real-time polymerase chain reaction for identification of dog meat in adulterated beef meatball using specific primer targeting on cytochrome-b for halal authentication. Int. J. Food. Prop. 23(1), 2231–2241. Rohman, A., Windarsih, A., Erwanto, Y. and Zakaria, Z., 2020b. Review on analytical methods for analysis of porcine gelatine in food and pharmaceutical products for halal authentication. Trends Food Sci. Technol. 101, 122–132. Salihah, N.T., Hossain, M.M., Lubis, H. and Ahmed, M.U. 2016. Trends and advances in food analysis by real-time polymerase chain reaction. J. Food. Sci. Technol. 53(5), 2196–2209. Sosiawan, A., Rohman, A. and Zain, N.M. 2020. The use of displacement loop mtDNA in halal forensic investigation in Indonesia. Res. J. Pharm. Technol. 13(3), 1069–1073. Sudjadi, Wardani, H.S., Sepminarti, T. and Rohman, A. 2016. Analysis of porcine gelatin DNA in a commercial capsule shell using real-time polymerase chain reaction for halal authentication. Int. J. Food. Prop. 19, 2127–2134. Tian, X., Wang, J., Ma, Z., Li, M., Wei, Z. and Díaz-Cruz, J.M. 2019. Combination of an E-nose and an E-tongue for adulteration detection of minced mutton mixed with pork. J. Food. Qual. 2019(1). Weedn, V.W., Gettings, K.B. and Podini, D.S. 2018. Identity testing, principles and applications of molecular diagnostics. Amsterdam, The Netherlands: Elsevier Inc. | ||
| How to Cite this Article |
| Pubmed Style Lestari LA, Aini WN, Laksitorini MD, Erwanto Y, Hastuti AAMB, Abidin MZ, Hidayah N, Budikafa MJ, Rohman A. Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study. Open Vet. J.. 2025; 15(1): 456-464. doi:10.5455/OVJ.2025.v15.i1.41 Web Style Lestari LA, Aini WN, Laksitorini MD, Erwanto Y, Hastuti AAMB, Abidin MZ, Hidayah N, Budikafa MJ, Rohman A. Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study. https://www.openveterinaryjournal.com/?mno=220693 [Access: September 04, 2025]. doi:10.5455/OVJ.2025.v15.i1.41 AMA (American Medical Association) Style Lestari LA, Aini WN, Laksitorini MD, Erwanto Y, Hastuti AAMB, Abidin MZ, Hidayah N, Budikafa MJ, Rohman A. Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study. Open Vet. J.. 2025; 15(1): 456-464. doi:10.5455/OVJ.2025.v15.i1.41 Vancouver/ICMJE Style Lestari LA, Aini WN, Laksitorini MD, Erwanto Y, Hastuti AAMB, Abidin MZ, Hidayah N, Budikafa MJ, Rohman A. Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study. Open Vet. J.. (2025), [cited September 04, 2025]; 15(1): 456-464. doi:10.5455/OVJ.2025.v15.i1.41 Harvard Style Lestari, L. A., Aini, . W. N., Laksitorini, . M. D., Erwanto, . Y., Hastuti, . A. A. M. B., Abidin, . M. Z., Hidayah, . N., Budikafa, . M. J. & Rohman, . A. (2025) Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study. Open Vet. J., 15 (1), 456-464. doi:10.5455/OVJ.2025.v15.i1.41 Turabian Style Lestari, Lily Arsanti, Wan Nurul Aini, Marlyn Dian Laksitorini, Yuny Erwanto, Agustina Ari Murti Budi Hastuti, Mohammad Zainal Abidin, Nurulia Hidayah, Muhammad Jefriyanto Budikafa, and Abdul Rohman. 2025. Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study. Open Veterinary Journal, 15 (1), 456-464. doi:10.5455/OVJ.2025.v15.i1.41 Chicago Style Lestari, Lily Arsanti, Wan Nurul Aini, Marlyn Dian Laksitorini, Yuny Erwanto, Agustina Ari Murti Budi Hastuti, Mohammad Zainal Abidin, Nurulia Hidayah, Muhammad Jefriyanto Budikafa, and Abdul Rohman. "Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study." Open Veterinary Journal 15 (2025), 456-464. doi:10.5455/OVJ.2025.v15.i1.41 MLA (The Modern Language Association) Style Lestari, Lily Arsanti, Wan Nurul Aini, Marlyn Dian Laksitorini, Yuny Erwanto, Agustina Ari Murti Budi Hastuti, Mohammad Zainal Abidin, Nurulia Hidayah, Muhammad Jefriyanto Budikafa, and Abdul Rohman. "Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study." Open Veterinary Journal 15.1 (2025), 456-464. Print. doi:10.5455/OVJ.2025.v15.i1.41 APA (American Psychological Association) Style Lestari, L. A., Aini, . W. N., Laksitorini, . M. D., Erwanto, . Y., Hastuti, . A. A. M. B., Abidin, . M. Z., Hidayah, . N., Budikafa, . M. J. & Rohman, . A. (2025) Application of real-time PCR for analysis canine meat (Canis lupus familiaris) in goat’s satay for halal authentication study. Open Veterinary Journal, 15 (1), 456-464. doi:10.5455/OVJ.2025.v15.i1.41 |