| Research Article | ||
Open Vet. J.. 2025; 15(1): 139-150 Open Veterinary Journal, (2025), Vol. 15(1): 139-150 Research Article First molecular detection of Eimeria spp. in domestic goats from Java Island, IndonesiaDias Aprita Dewi1,2, Yudhi Ratna Nugraheni1, Aan Awaludin1,3, Vika Ichsania Ninditya1, Dwi Priyowidodo1, Raden Wisnu Nurcahyo1, Fitrine Ekawasti4 and Joko Prastowo1,*1Department of Parasitology, Faculty of Veterinary Medicine, Universitas Gadjah Mada, Yogyakarta, Indonesia 2Departmenf of Animal Science, Politeknik Pembangunan Pertanian Yogyakarta Magelang, Magelang, Indonesia 3Department of Animal Science, Politeknik Negeri Jember, Jember, Indonesia 4Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia *Corresponding Author: Joko Prastowo. Department of Parasitology, Faculty of Veterinary Medicine, Universitas Gadjah Mada, Yogyakarta, Indonesia. Email: joko2465 [at] ugm.ac.id Submitted: 18/09/2024 Accepted: 18/12/2024 Published: 31/01/2025 © 2025 Open Veterinary Journal
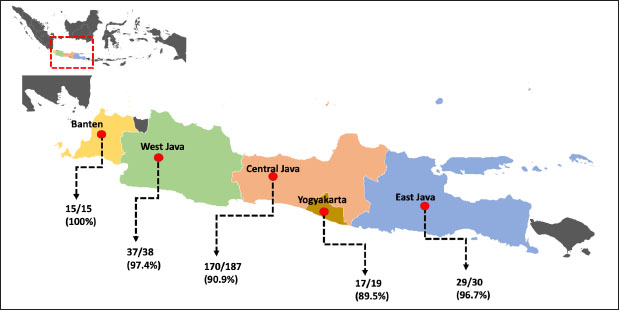
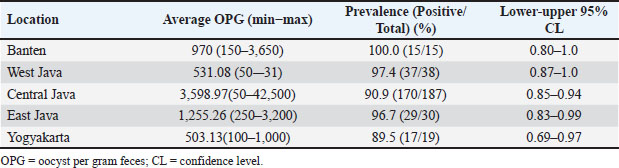
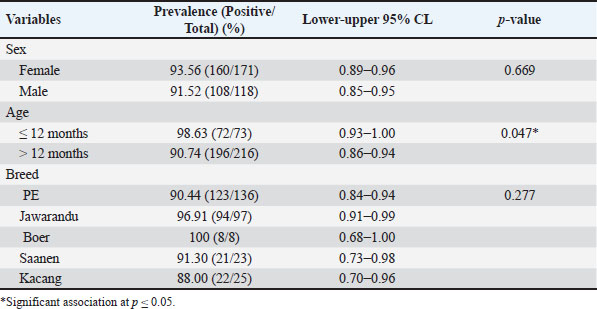
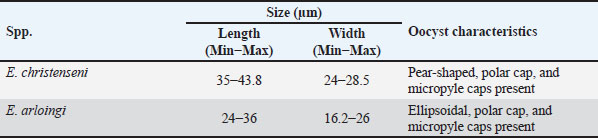
AbstractBackground: Coccidiosis caused by Eimeria species (spp.) is a significant global health concern in goats leading to gastrointestinal illness. This condition causes clinical manifestations, including weight loss and diarrhea, resulting in worldwide economic losses. Subclinical symptoms can manifest during Eimeria infection. Neglecting this disease can lead to severe morbidity and mortality. Therefore, addressing caprine coccidiosis is imperative. Aim: This study aimed to determine the prevalence and molecular identification related to the natural infection of Eimeria spp. in domestic goats originating from Java Island, Indonesia. Methods: In total, fecal samples from 289 domestic goats were obtained across five provinces on Java Island, Indonesia: East Java, Central Java, D. I. Yogyakarta, West Java, and Banten. Morphological examinations were performed using the modified Whitlock method and saturated sugar flotation. Molecular assays targeting the 18S ribosomal ribonucleic acid have been employed for spp.-specific confirmation. Statistical analysis was performed using the Wilson binomial proportion and chi-square methods implemented in the online software. Results: A total of 92.7% (268/289) of fecal samples tested positive for Eimeria spp. Phylogenetic tree analysis demonstrated that Eimeria christenseni and Eimeria arloingi closely resembled the reference sequences from China, Australia, and other countries. Conclusion: This study identified E. christenseni and E. arloingi as the goat-infecting spp. of Eimeria present on Java Island. The specific and accurate molecular identification conducted in this study will contribute to improved coccidiosis control and the development of effectiveness. Keywords: Eimera, Goat, Indonesia, Molecular identification, 18S rRNA. IntroductionSmall ruminants play a significant role in sustainable agriculture in developing countries and provide numerous socioeconomic advantages (Anim-Jnr et al., 2023). Goats are considered one of the most promising sources of meat and dairy production in Indonesia. Indonesia is a tropical country that has a total goat population of around 18.5 million with various local breeds including Peranakan Ettawa, Jawarandu, Saanen, Boer, and Kacang (BPS, 2022). Eimeria spp. are gastrointestinal protozoans belonging to the apicomplexan group (Underwood et al., 2015). These protozoans infect the digestive tract, causing coccidiosis. Coccidiosis has high morbidity and mortality rates as the condition causes impaired nutrient absorption, which results in decreased quality of life and productivity (Engidaw et al., 2015). Several Eimeria species (spp.) have been identified in small ruminants, with those frequently infecting goats including Eimeria caprina, Eimeria ninakohlyakimovae, Eimeria jolchijevi, Eimeria apsheronica, Eimeria christenseni, Eimeria caprovina, Eimeria alijevi, Eimeria hirci, and Eimeria arloingi (Bawm et al., 2020). Among these, E. arloingi, E. ninakohlyakimovae, and E. christenseni are highly pathogenic and are often associated with significant mortality in goats, especially those under one year of age (Khodakaram-Tafti et al., 2013; Roh et al., 2023). Clinical symptoms are insufficient for the diagnosis of coccidiosis. This could be misinterpreted as a clinical manifestation of other intestinal disorders. The diagnosis of coccidiosis typically relies on the morphological characteristics of oocysts, including host specificity, aspects of the life cycle, sites of infection, pathogenicity, antigenicity, nucleotide sequencing, clinical symptoms in the animal, and typical macroscopic lesions at necropsy (Berto et al., 2014). Information on the morphological characteristics of Eimeria in Indonesia remains limited; therefore, the specific Eimeria spp. present in Indonesia remains unclear. When compared, those spp. identified across different regions of other countries have slightly different morphological and structural variations (Gondipon and Malaka, 2021). The identification of different spp. has traditionally relied on the microscopic examination of oocysts, which involves biological assessments, including the size and morphological characteristics of oocysts, absence or presence of residual oocysts, sporulation period, prepatent period, micropyle cap, and site of colonization (Bawm et al., 2020). This method is commonly employed to identify coccidia in goats, particularly in Indonesia (Pamungkas et al., 2021; Pertiwi et al., 2023). However, due to the similar morphological characteristics of some spp., relying on physical traits alone is not sufficient for accurately identifying Eimeria spp. with nearly identical shapes and sizes (morphological resemblance) (Haug et al., 2007). To address this limitation, polymerase chain reaction (PCR) has been used to accurately and rapidly identify Eimeria spp. (Jordan et al., 2018). Molecular assays are considered precise methods for identifying Eimeria spp. due to their high sensitivity and reliability (Al-Habsi et al., 2017). Deoxyribonucleic acid (DNA) sequences are known to be more consistent than morphological characteristics. In the present study, identification and phylogenetic analyses were performed using ribosomal DNA (rDNA). rDNA in eukaryotic cells consists of tandem arrays containing several hundred to thousand repetitions in each arrangement (Hillis and Dixon, 2011). The 18S ribosomal ribonucleic acid (rRNA) gene is frequently utilized as a molecular marker for genetic analysis. Furthermore, 18S rRNA gene sequences have been used to delineate the inter- and intraspecies phylogenetic relationships among various Eimeria isolates (Verma et al., 2017). Despite limited knowledge of the genetic characteristics of Eimeria spp. in small ruminants, molecular biology methods have the potential for appropriate classification. This study aimed to bridge this gap by isolating genomic DNA from purified oocysts in goats, followed by amplification and sequencing of the 18S rRNA loci. Subsequent phylogenetic analyses of Eimeria using the obtained sequences could contribute to the identification of spp., the epidemiology of molecular biology, and phylogenetic analysis. Materials and MethodsStudy design and sample collectionThis cross-sectional study was conducted to determine the prevalence of coccidiosis in domestic goats across five provinces on Java Island, Indonesia: East Java, Central Java, D.I. Yogyakarta, West Java, and Banten. In this study, 289 fecal samples were obtained from five provinces. In Banten Province, 15 samples were collected from a single farm, with 15 samples obtained from each farm. A total of 38 samples were collected from two farms located in the West Java Province, with each farm providing between 18 and 20 samples. Central Java Province included 187 samples collected from 11 farms, with each farm providing 18–20 samples. In East Java, 30 samples were gathered from two farms, with 15 samples being obtained from each farm. In D. I. Yogyakarta, 19 samples were collected from two farms, with each farm contributing between nine and 10 samples. The minimum sample size was calculated using the formula n=4PQ/L2 (Martin et al., 1987). The calculation was based on a predicted prevalence of 82%, 95% confidence level, and 5% statistical error (de Macedo et al., 2020). A total of 289 fecal samples of approximately 15 g (± 15 g) each were obtained at random. Fresh samples were collected directly from the rectum and stored in plastic containers. They were then transported to the Department of Parasitology, Faculty of Veterinary Medicine, Universitas Gadjah Mada, for further analysis. Identification of Eimeria spp. by morphological examinationSaturated sugar flotation methodFeces (1 g) from each animal were dissolved in 9 ml of water and left for a few minutes until soft, crushed, stirred homogeneously, and filtered through a steel mesh. The fecal solutions were placed in centrifuge tubes and centrifugated at 1,500 rpm for 5 minutes. The supernatant was discarded, and the tube was then filled with 10 ml saturated sugar float solution with a selected gravity of 1.2 (e.g., 100 g of sugar was added to 120 ml of distilled water). The mixture was mixed until homogenous and then centrifuged for 5 minutes at 1,500 rpm. The centrifuge tube was gently removed from the machine and positioned upright on a test-tube rack. The floating fluid was added dropwise using a Pasteur pipette until the surface formed a convex shape, ensuring that no excess fluid spilled over. After waiting for 5 minutes for the oocysts to float to the surface, a cover glass was placed on the surface of the liquid and attached to the object glass. Oocysts were detected based on their shape and size under a microscope at 400× magnification (Mohamaden et al., 2018; Efendi et al., 2019). McMaster method (Gordon and Whitlock technique)This approach was used to count coccidian oocysts. Feces from each animal (up to 3 g) were dissolved in water (up to 17 ml for a few minutes before being crushed and mixed with a saturated salt solution (40 ml to allow the coccidia oocysts to float). Afterward, while stirring, the stool solution was collected containing the filter and was transferred to the Whitlock chamber (Gordon and Whitlock, 1939). Coccidia oocysts were assessed using a microscope and multiplied by 40. The number of oocysts was calculated as oocysts per gram (OPG) based on microscopic observations. According to Gondipon and Malaka (2021), the degree of infection could be classified as light (50–799), medium (800–1,200), or high (> 1,200). Purification of Eimeria spp. oocystsEimeria spp. oocysts were purified from goat feces by filtration and centrifugation using a saturated sugar solution to obtain purified oocysts (Kawahara et al., 2010). Additionally, Eimeria oocysts were isolated from the remaining feces (approximately 5–10 g). The feces were then diluted with distilled water and filtered through a steel mesh or gauze. After centrifugation at 800 rpm for 5 minutes, the sugar solution was added to the sample, which was then centrifuged at 1,200 rpm for 10 minutes. Eimeria oocysts floating on the surface were collected using a Pasteur pipette and rinsed with distilled water. Purified oocysts were suspended in 1–2 ml distilled water (Hastutiek et al., 2022). Sporulation of Eimeria spp. oocystsThe purified oocysts were sporulated in 2.5% potassium dichromate solution at 26°C–33°C for 5 d. A microscope with 400× magnification was employed to identify oocysts and sporocysts. Morphological analysisMorphological analysis was performed using the identification key developed by Eckert et al. (1995). Sporulation oocysts were observed under a microscope at 400× magnification, and morphological features such as size, shape, and the presence of specific structures including the micropyle and polar cap were evaluated (Mohamaden et al., 2018; Efendi et al., 2019). Identification of Eimeria spp. by molecular examinationDNA extractionBefore DNA extraction, purified oocyst samples from goat feces were frozen and thawed to break the oocyst walls mechanically, thereby releasing the genomic DNA (Lassen and Järvis, 2009; Gadelhaq et al., 2015). The freeze-thawing process was carried out by storing the sample in a freezer at −20oC for 15 minutes, followed by transferring it to a heater at 60oC for 15 minutes until it melted. The sample was then spun down, and this cycle was repeated five times (Matsubayashi et al., 2009). DNA was extracted using the Geneaid GT100 Genomic DNA Mini Kit (Geneaid, Taiwan). Amplification and sequencing of 18S rRNAEimeria spp. were identified using PCR targeting the 18S rRNA region. Partial sequences of the 18S rRNA gene were amplified with 630 bp using forward primer 5′-TACCCAATGAAAACAGTTT-3′ and reverse primer 5′-CAGGAGAAGCCAA GGTAGG-3′ according to the previous study conducted by Jinneman et al. (1999). The PCR reaction was performed in a 25 µl volume, which included 0.75 µl forward and reverse primers (10 µM), 12.5 µl of 2× Buffer (KOD Fx Neo), 0.5µl of Taq Polymerase, DNA template (2 µl), and nuclease-free water. Positive samples used in this study were obtained from samples with very high parasitemia levels, amplified by PCR, and confirmed as Eimeria by Sanger sequencing. Sterile DNA/RNA-free water was used as the negative control. The PCR condition was set with an initial denaturation step (2 minutes at 94°C), followed by 35 cycles of denaturation (10 s at 98°C), annealing (30 s at 51°C), and extension (20 s at 68°C). A final extension step was performed at 68°C for 5 minutes. The PCR products were separated by gel electrophoresis on a 1.5% (w/v) agarose gel in 1× TAE buffer, run at 120 V for 30 minutes, and visualized using an ultraviolet transilluminator. Positive PCR samples were subjected to Sanger sequencing at the Penelitian dan Pengujian Terpadu UGM Laboratorium. Sequencing findings, represented by the nucleotide base sequence in the chromatogram file, were analyzed. Sequences analysisSequences from this study and GenBank were aligned using ClustalW in BioEdit version 7 (Hall, 1999). After alignment, sequences were compared with GenBank data using BLASTN software. Phylogenetic trees were created using IQtree version 1.6. The trees were visualized using FigTree v1.4.4 (available at http://tree.bio.ed.ac.uk/software/figtree/). All sequences obtained in this study were deposited in GenBank with accession numbers PQ222707–PQ222721. Statistical analysisThe proportion was analyzed using Wilson’s method with confidence intervals. The chi-square test was employed to investigate the association between coccidia infection, sex, and age. All data were evaluated using a statistical analysis tool incorporated into an epidemiological calculator which is free and accessible at https://epitools.-ausvet.com.au. The prevalence of samples from each province was statistically analyzed using the Binomial (Clopper–Pearson) “exact” method based on the beta distribution. Ethical approvalThe current study was conceived and performed in accordance with the animal welfare regulations issued by the Research Ethics Committee of the Faculty of Veterinary Medicine, Universitas Gadjah Mada, Indonesia. ResultsA total of 289 domestic goat fecal samples were obtained from five provinces on Java Island, Indonesia, including East Java, Central Java, D. I. Yogyakarta, West Java, and Banten, and evaluated for coccidia oocysts using the saturated sugar flotation method for qualitative analysis and the Gordon–Whitlock method for quantitative analysis. The results demonstrated that 92.7% (268/289) of the fecal samples tested positive for Eimeria spp. In Banten Province, the OPG ranged from 150 to 3,650, with all samples (100%) testing positive for coccidia infection. Similarly, in West Java Province, the OPG ranged from 50 to 531, with a high proportion of positive samples at 97.4% (37/38). In Central Java Province, the OPG ranged from 50 to 42,500, with 90.9% (170/187) of the samples testing positive for coccidia. Furthermore, in East Java Province, the OPG ranged from 250 to 3,200, with 96.7% (29/30) of the samples testing positive. In D. I. Yogyakarta Province, the OPG ranged from 100 to 1,000, with 89.5% (17/19) of the samples testing positive (Fig. 1). Table 1 presents the evaluation results, demonstrating the range of OPG in feces, the proportion of positive samples, and corresponding confidence intervals (95%) for each location. Variables including sex, age, and breed were associated with the univariate analysis as risk factors for infection in goats. According to an epidemiological study, one of the three variables was significantly related to Eimeria infection, namely, age, while the others were not correlated (Table 2). Eimeria spp. identification by morphologyMorphological features were assessed according to the micropyle, polar cap, and oocyst size and shape. The length and width of the oocysts were also evaluated (Christensen, 1938). In this study, we identified the oocysts of E. christenseni and E. arloingi based on microscopic observations of sporulated oocysts. The specific characteristics of each spp. are described in detail in Table 3. Furthermore, Figure 2 illustrates the goat coccidia used in this study. Molecular characteristic of Eimeria spp.The 18S rRNA gene was used for PCR after extracting DNA from 18 fecal samples that were morphologically positive for Eimeria spp. All samples produced positive bands, and the PCR product was approximately 630 bp. No amplification products were observed in the negative controls. The 18 PCR-positive samples were sequenced. Unfortunately, three samples were deemed to have low-quality chromatography results. Thus, we excluded these sequences, leaving 15 sequences for molecular and phylogenetic analyses. To determine the spp. of Eimeria oocyst using molecular method, 18S rRNA gene sequences were analyzed using BLAST against an extensive non-redundant nucleotide database. After removing low-quality sequences from 18 sequences, 15 high-quality sequences were selected for molecular investigation. Additionally, analysis performed using the BLASTN program employing the 18S rRNA sequences obtained from the sample revealed that they were 100% identical to E. christenseni (accession no. KX845684, OM328173, OM328172, OM328171), meanwhile, Eimeria 3 was 99.4%–99.8% identical to E. arloingi (accession no. OM328169, ON259586).
Fig. 1. Map depicting sampling site collection in this study. Prevalence rate samples are presented under the arrow, respectively. Table 1. OPG of feces assessed by the Whitlock method.
Table 2. Univariate analysis of the risk factors associated with Eimeria infection in goats from Java Island.
Table 3. Morphological characteristics of Eimeria oocysts obtained from goat feces in this study.
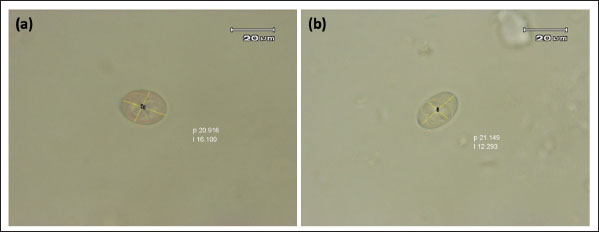
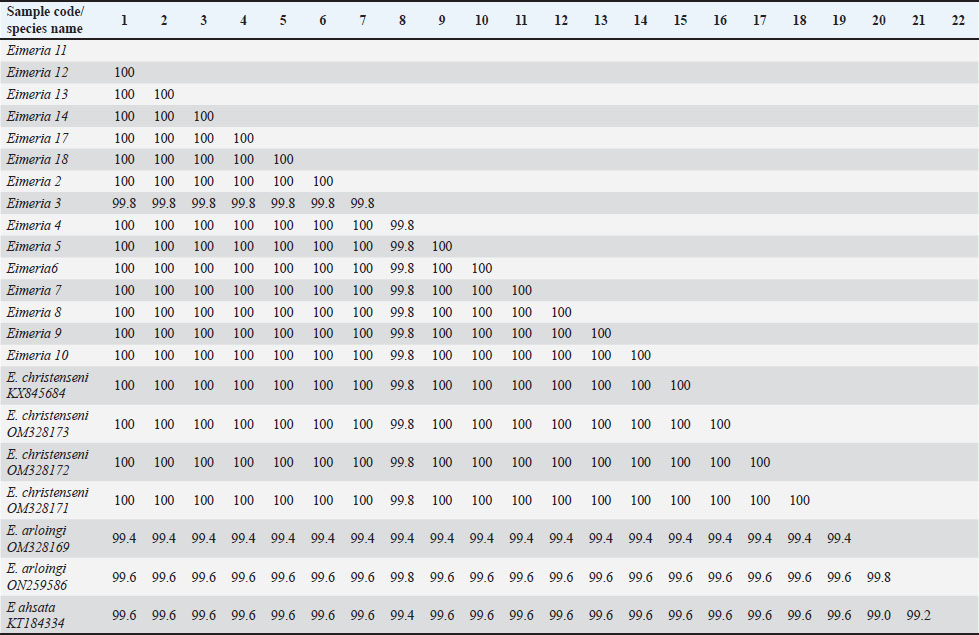
Fig. 2. Sporulated E. christenseni (a) and E. arloingi (b). According to the results of a comparative study of the sequences, 99.4% to 100% of the Eimeria spp. from goat samples were identified and were similar to E. christenseni and E. arloingi. Additionally, the sequence similarities between Eimeria spp. from goats and Eimeria ahsata from sheep ranged from 99.0% to 99.6% (Table 4). A maximum likelihood phylogenetic tree was constructed using the 18S rRNA gene, incorporating Eimeria isolates and reference Eimeria spp. from GenBank. BLAST analysis was performed to match the reference sequences from GenBank with those of the Eimeria isolates. Our findings revealed a strong correlation between Eimeria isolates and Eimeria spp. from Myanmar, China, Iran, and Australia (Fig. 3). Table 4. Pairwise identities (%) among the 18S rRNA sequences of Eimeria samples with E. christenseni, E. arloingi, and E. ahsata.
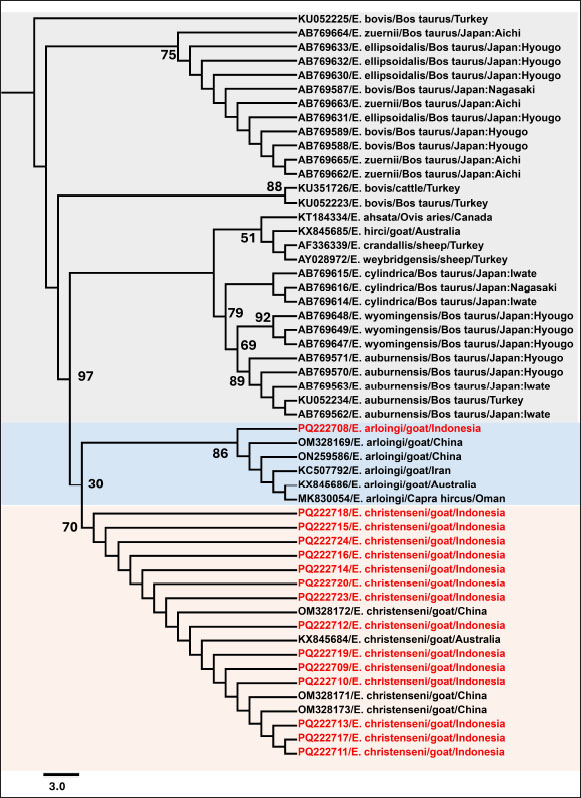
A close association between the isolates of Eimeria spp. was revealed by the phylogenetic tree analysis with high sequence homology to reference strains. The 18S rRNA sequences from China and Australia were completely similar to E. christenseni (accession numbers: KX845684, OM328173, OM328172, and OM328171), forming a distinct clade. Additionally, one sequence clustered with E. arloingi (accession numbers OM328169 and ON259586) from China and displayed 100% sequence homology.
Fig. 3. Maximum likelihood tree of the partial 18S rRNA gene for approximately 650 bp. The phylogenetic tree was generated by Bootstrap 1,000, and the best-fit model K2P+I was chosen according to the Bayesian Information Criterion (BIC). Eimeria bovis was used as an outgroup. Bootstrap values are indicated at each node. DiscussionGoat farming is an important aspect of livestock farming in Indonesia (Tenrisanna and Kasim, 2021). Currently, coccidiosis significantly impacts the production traits of goats, such as milk and meat, leading to substantial economic losses worldwide. This has become a major concern hindering the healthy growth of the goat farming industry and poses a serious threat to the health and productivity of livestock, particularly young goats. Understanding the incidence of Eimeria in a specific region to implement effective control measures and minimize the economic losses associated with the disease is crucial (Ruiz et al., 2012). Sixteen Eimeria spp. have been documented to affect goats globally, although the precise number of valid spp. varies depending on the study (Abo-Shehada and Abo-Farieha, 2003). Currently, the practical method for identifying spp. within goat coccidia is through morphological observation of oocysts (Khodakaram-Tafti et al., 2013). Nonetheless, this approach may prove unreliable due to potential overlap in the quantitative and qualitative morphological characteristics of oocyst between spp. and within spp. of Eimeria. To address the limitations of traditional methods, molecular techniques have recently been demonstrated to be effective in identifying and classifying Eimeria (Matsubayashi et al., 2005; Kawahara et al., 2010; Takeo et al., 2014). Identifying spp. in Eimeria co-infections is difficult because of their morphological similarities, which complicates their differentiation under a microscope. The presence of overlapping life cycle stages complicates identification, as various spp. may exhibit similar characteristics during certain developmental phases. Mixed infections further complicate the evaluation of the contribution of each spp. to the disease severity. These issues underscore the need to integrate multiple diagnostic techniques, including molecular assays, to identify cases of co-infection reliably. In Indonesia, morphological and molecular studies focusing on Eimeria spp. in goats are limited. This study aimed to identify the prevalence and composition of Eimeria spp. causing coccidiosis in domestic goats in Java, Indonesia, using morphological examination and molecular methods with 18S rRNA sequences. This could contribute to the understanding of the biology, molecular epidemiology, and population diversity of goat Eimeria spp. Additionally, the analysis will aid in supporting host adaptation of Eimeria spp. in various animals, including goats, sheep, and cattle. This study revealed that two Eimeria spp. have adapted to goats in different provinces across Java Island. The two different spp. suggests that Eimeria responds to regional immune systems and farming practices. High infection rates may favor organisms with enhanced infection or treatment resistance. The analysis of fecal samples using saturated sugar flotation and Whitlock techniques has enabled the detection and measurement of Eimeria oocysts. These methods are widely used because of their ease and reliability in identifying coccidian infections (Khodakaram-Tafti et al., 2013). Our study identified suspected cases of E. christenseni and E. arloingi, which are known to have high pathogenicity and are often fatal to goats, especially those under one year of age (Roh et al., 2023). The sporulation process of Eimeria oocysts in potassium dichromate facilitates morphological characterization and spp. identification (Gong et al., 2021). Mature sporulated oocysts were subjected to microscopic examination, where morphological features including shape, size, and the presence of specific structures such as the micropyle and polar cap were evaluated (Stotish et al., 1978). Based on these characteristics, E. christenseni and E. arloingi were identified and their morphological differences were noted. The current study demonstrated a moderately high prevalence of Eimeria spp. in goats (92.7%, 268/289) on Java Island, Indonesia, with varying prevalence rates in each province. Certain regions exhibit higher prevalence rates than those noted in others. Confidence intervals estimate the precision of the proportion of positive samples at each location. The prevalence of E. christenseni and E. arloingi is widely recognized as the most pathogenic coccidia spp. affecting goats globally (Roh et al., 2023). A morphological study using microscopic examination of goats in the arid tropical regions of Africa, such as Kenya (Kanyari et al., 2009), Somalia (Harmaya et al., 2020), and Nigeria, revealed a significant prevalence of Eimeria spp. infections. Notably, E. christenseni (60%) and E. arloingi (64%–80%) were identified as the most prevalent spp. Comparative research from Asian countries, such as Vietnam, India, Myanmar, and China, also demonstrated a low to a high prevalence of E. christenseni (63%–78%) and E. arloingi (21%–73%) (Bawm et al., 2020; De et al., 2021; Diao et al., 2022; Nguyen-Ho-Bao et al., 2022). The absence of statistically significant differences in coccidia infection rates between the two variables (sex and breed) suggests that infection may occur in goats indiscriminately based on these factors. In terms of sex, females (93.56%) had a non-significantly higher infection rate than the rate in males (91.52%). Similar results for the prevalence of Eimeria spp. infection between sexes have been reported previously by Dabasa et al. (2017) attributing to the similar management systems used to maintain both sexes, which provides an equal likelihood of infection. The animals in this study demonstrated a non-significant difference in the prevalence of Eimeria spp. Infections. Moreover, Souza et al. (2015) reported that Eimeria infection can affect small ruminants of all breeds. All breeds in the region experience the same hot and humid climate, which is conducive to the sporulation of coccidian oocysts. Rehman et al. (2011) reported that different breeds had no genetic influence on the prevalence of Eimeria. According to Ahmed (2011), natural resistance-associated macrophage protein one is associated with factors affecting the susceptibility of goats to developing coccidia; however, no breed effect was detected. Young goats (age ≤ 12 months) (98.63%) were more affected than adult goats (90.74%). Age is an important risk factor for Eimeria infections in sheep (Carneiro et al., 2022). However, another study reported that animals of different age groups were similarly exposed to Eimeria spp. (de Macedo et al., 2020). Adult animals can develop parasite immunity through repeated challenges and can eliminate the parasite before the development of the infection (López-Osorio et al., 2020). Consequently, coccidiosis is considered a self-limiting disease. Additionally, the disease gradually increases in frequency and severity, peaking just before weaning and then declining in adults. This finding underscores the importance of implementing preventive measures across all goat age groups (Bawm and Htun, 2023). Identifying E. christenseni and E. arloingi in this study adds to the existing knowledge on Eimeria spp. diversity in Java, Indonesia. These spp. are highly pathogenic and can produce severe clinical manifestations in goats, leading to significant economic losses in the livestock industry (Dai et al., 2006; Guedes et al ., 2024). Overall, these findings underscore the widespread distribution of coccidia infection among domestic goats on Java, highlighting the need for effective control measures to reduce the effects of disease on the productivity and health of livestock. Therefore, continued surveillance and research efforts are warranted to monitor changes in the prevalence and distribution of Eimeria spp. and develop targeted control strategies. Identifying Eimeria spp. in goats has historically relied on morphological analysis. In the present study, we employed molecular techniques based on PCR amplification of the parasite to confirm our findings. This study employed 18S rRNA gene sequencing to confirm Eimeria at the spp. level. The 18S rRNA gene has been extensively used as a molecular marker in phylogenetic analyses due to its high specificity. These sequences have proven effective in differentiating between various closely related Eimeria spp. (Matsubayashi et al., 2005; Pyziel et al., 2020). Recently, the 18S rRNA gene was used as a target to distinguish closely related spp. of Eimeria and conduct phylogenetic analyses (Liang et al., 2022). In addition, analysis of nucleotide sequences using the maximum likelihood method verified that Eimeria isolated from goats in this study were genetically grouped with the previously described E. christenseni and E. arloingi. Phylogenetic analysis revealed a comprehensive overview, indicating that E. christenseni and E. arloingi formed distinct taxonomic groups, separate from the other Eimeria spp. in cattle and sheep. The current study contradicts the findings of Liang et al. (2022). Their study demonstrated that phylogenetic analysis based on 18S rDNA indicated E. christenseni was the most closely related to ovine Eimeria, followed by E. ellipsoidaliss, E. zuernii, and E. bovis from cattle. Notably, E. arloingi demonstrated 99% similarity to E. ahsata based on 18S rRNA (Khodakaram-Tafti et al., 2013). Recognizing certain limitations of this study is imperative. The lack of comprehensive genetic sequences for essential Eimeria spp. in GenBank for the target gene under study makes it challenging to perform a comparative analysis of coccidia across other goat spp. This gap reduces the accuracy of spp. identification in mixed infections, impede evaluation of genetic diversity, and complicate the development of effective molecular diagnostic tools. The precise diagnosis of the causal agent of coccidia infection is critical for gaining a better understanding of the parasite’s biology and life cycle. Identifying spp. in Eimeria co-infections is difficult because of their morphological similarities, which complicates their differentiation under a microscope. The presence of overlapping life cycle stages complicates identification, as various spp. may exhibit similar characteristics during certain developmental phases. Mixed infections further complicate the evaluation of the contribution of each spp. to the disease severity. These issues underscore the need to integrate multiple diagnostic techniques, including molecular assays, to reliably identify cases of co-infection. This study revealed significant differences in Eimeria infection rates among various provinces of Java, Indonesia. This study identified that E. christenseni and E. arloingi were the primary spp. causing caprine coccidiosis. Valuable molecular data for E. christenseni and E. arloingi were obtained, which can be utilized for future molecular research. Therefore, the sequence obtained in this study will contribute to expanding our knowledge of the genetic diversity and geographical distribution of Eimeria spp. that infect small ruminants worldwide. Further research on this topic is necessary in regions of Indonesia beyond Java Island, with a significant focus on the molecular identification and characterization of the Eimeria spp. These findings offer important baseline data that can help develop strategies for managing the disease in Indonesia. In conclusion, this study provides information regarding the types of Eimeria spp. in goats in Java, Indonesia, along with their occurrence levels. Additionally, E. christenseni and E. arloingi were among the spp. identified. These results highlight the epidemiology of coccidiosis in the region and the importance of disease control in goat farms to anticipate economic losses associated with this disease in Java, Indonesia. AcknowledgmentsDias Aprita Dewi received a scholarship from the Agricultural Extension and Human Resource Development Agency, Ministry of Agriculture, while Aan Awaludin was supported by the Center for Higher Education Funding (BPPT) through the Indonesian Education Scholarship/Beasiswa Pendidikan Indonesia program and the Indonesia Endowment Fund for Education (Agency/Lembaga Pengelola Dana Pendidikan). Conflicts of interestThe authors declare they have no conflict of interest. FundingThis research was supported by a Final Project Recognition/Rekognisi Tugas Akhir (RTA) Grant, Universitas Gadjah Mada (grant number 5286/UN1). P.I/PT.01.03/2024. Authors’ contributionsDAD: sample collection, morphological examination, Whitlock examination, DNA sequence analysis, writing the original manuscript; YRN: sample collection, editing sequencing results, molecular data analysis, manuscript editing; AA: sample collection, oocyst measurement and quantification; VIN: DNA extraction, DNA purification, manuscript editing; DP: PCR performance, statistical analysis, morphological examination; RWN: PCR performance, PCR analysis, manuscript editing; FE: morphological data analysis, quantification of OPG in feces; JP: morphological examination, manuscript editing, funding acquisition. Data availabilityAll data supporting the findings of this study are available within the manuscript. ReferencesAbo-Shehada, M.N. and Abo-Farieha, H.A. 2003. Prevalence of Eimeria species among goats in northern Jordan. Small. Rumin. Res. 49(2), 109–113. Ahmed, Y. 2011. Factors affecting susceptibility of goats to Coccidia. Greensboro, NC: North Carolina Agriculture and Technical State University. Al-Habsi, K., Yang, R., Ryan, U., Miller, D.W. and Jacobson, C. 2017. Morphological and molecular characterization of three Eimeria species from captured rangeland goats in Western Australia. Vet. Parasitol. Reg. Stud. Rep. 9, 75–83. Anim-Jnr, A., Sasu, P., Bosch, C., Mabiki, F.P., Frimpong, Y.O., Emmambux, M.N. and Greathead, H.M.R. 2023. Sustainable small ruminant production in low- and middle-income African countries: harnessing the potential of agroecology. Sustainability 15(21), 15326. Bawm, S. and Htun, L.L. 2021. Management and control of Eimeria infection in goats. In Goat science-environment, health and economy. Eds., Kukovics, S. London: Intech Open. pp: 1–14. Bawm, S., Win, T.Z.B., Win, S.Y., Htun, L.L., Nakao, R. and Katakura, K. 2020. First detection of Eimeria species in Myanmar domestic goats with both microscopic and molecular methods. Parasite 27(38), 1–7. Berto, B.P., Mcintosh, D. and Lopes, C.W.G. 2014. Studies on coccidian oocysts (Apicomplexa: Eucoccidiorida). Rev. Bras. Parasitol. Vet. 23(1), 1–15. BPS. 2022. Populasi Kambing menurut Provinsi (Ekor), 2021–2023. Jakarta, Indonesia: BPS Indonesia. Carneiro, P.G., Sasse, J.P., Silva, A.C.D.S., de Seixas, M., Paschoal, A.T.P., Minutti, A.F., Martins, T.A., Cardim, S.T., de Rodrigues, F.S., de Barros, L.D. and Garcia, J.L. 2022. Prevalence and risk factors of Eimeria spp. natural infection in sheep from northern Paraná, Brazil. Rev. Bras. Parasitol. Vet. 31(1), 1–7 . Christensen, J.F. 1938. Species differentiation in the Coccidia from the domestic sheep. J. Parasitol. 24(5), 453. Dabasa, G., Shanko, T., Zewdei, W., Jilo, K., Gurmesa, G. and Abdela, N. 2017. Prevalence of small ruminant gastrointestinal parasites infections and associated risk factors in selected districts of Bale zone, south eastern Ethiopia. J. Parasitol. Vector Biol. 9(6), 81–88 . Dai, Y.B., Liu, X.Y., Liu, M. and Tao, J.P. 2006. Pathogenic effects of the coccidium Eimeria ninakohlyakimovae in goats. Vet. Res. Commun. 30(2), 149–160. de Macedo, L.O., Bezerra-Santos, M.A., de Mendonça, C.L., Alves, L.C., Ramos, R.A.N. and de Carvalho, G.A. 2020. Prevalence and risk factors associated with infection by Eimeria spp. in goats and sheep in Northeastern Brazil. J. Parasit. Dis. 44(3), 607–612. De, A.K., Perumal, P., Muniswamy, K., Zamir Ahmed, S.K., Kundu, A., Sunder, J., Alyethodi, R.R., Ravi, S.K. and Bhattacharya, D. 2021. Prevalence of coccidiosis in Andaman local goat and its metaphylaxis in tropical island ecosystem. Indian J. Anim. Res. 91(6), 438–442 . Diao, N.C., Zhao, B., Chen, Y., Wang, Q., Chen, Z.Y., Yang, Y., Sun, Y.H., Shi, J.F., Li, J.M., Shi, K., Gong, Q.L. and Du, R. 2022. Prevalence of Eimeria spp. among goats in China: a systematic review and meta-analysis. Front. Cell. Infect. Microbiol. 12, 1–11. Eckert J., Braun R., Shirley, M.W. and Coudert P. 1995. Biotechnology: guidelines on technique in coccidiosis research. Luxembourg, Europe: ECSC-EC-EAEC. pp: 25–39 Efendi, W.N., Suwanti, L.T., Samik, A., Hastutiek, P., Mufasirin, and Kusnoto. 2019. Prevalensi dan Identifikasi Protozoa Saluran Pencernaan pada Kambing di Kecamatan Labang Kabupaten Bangkalan. J. Parasite Sci. 3(2), 9–25. Engidaw, S., Anteneh, M. and Demis, C. 2015. Coccidiosis in small ruminants. Afr. J. Appl. Res. 7(6), 311–319 . Gadelhaq, S.M., Arafa, W.M. and Aboelhadid, S.M. 2015. Molecular characterization of Eimeria species naturally infecting Egyptian baldi chickens. Iran. J. Parasitol. 10(1), 87–95. Gondipon, R. and Malaka, R. 2021. Overview of coccidiosis in sheep: history of disease incidence in the world and life cycle. HAJAS. 3(1), 42–51. Gong, Z., Wei, H., Chang, F., Yin, H. and Cai, J. 2021. Sporulation rate and viability of Eimeria tenella oocysts stored in potassium sorbate solution. Parasitol. Res. 120(6), 2297–2301. Gordon H. and Whitlock H.V. 1939. A new technique for counting nematode eggs in sheep faeces. J. Council Sci. Ind. Res. 12, 50–52. Guedes, A.C., Conde-Felipe, M., Barba, E., Molina, J.M., del Carmen Muñoz, M., Ferrer, O., Martín, S., Hermosilla, C., Taubert, A. and Ruiz, A. 2024. Metaphylactic strategies using toltrazuril against coccidiosis in goat kids. Vet. Parasitol. 327, 110133. Hall, T.A. 1999. BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp. Ser. 41, 95–98. Harmaya, A., Zegeye, B., Alemu, S., Temesgen, W., Emiru, B., Amede, Y., Tigre, W., Feyera, T., Deressa, B., Teshome, D., Tessema, T., Kumsa, S., Naramo, M., Gupta, N., Singh, N., Sonkar, N., Njau, B.C., Scholtens, R.G., Kasali, O. and Kiros, Y. 2020. Study on prevalance of small ruminant Coccidiosis in and Around Harmaya, Eastern Haraghe Ethiopia study on prevalance of small ruminant Coccidiosis in. Eur. J. Biol. Res. 7(1), 1–5. Hastutiek, P., Lastuti, N.D.R., Suwanti, L.T., Sunarso, A., Suprihati, E., Kurniawati, D.A. and Matsubayashi, M. 2022. Coproparasitological examinations and molecular determination of Eimeria species in Madura cattle reared on Madura Island, Indonesia. Parasitol. Int. 86, 102478. Haug, A., Thebo, P. and Mattsson, J.G. 2007. A simplified protocol for molecular identification of Eimeria species in field samples. Vet. Parasitol. 146(1–2), 35–45. Hillis, D.M. and Dixon, M.T. 2011. Ribosomal DNA : molecular evolution and phylogenetic inference December 1991 ribosomal DNA : molecular evolution and phylogenetic inference. Rev. Lit. Arts Am. 66(4), 411–453. Jinneman, K.C., Wetherington, J.H., Hill, W.E., Omiescinski, C.J., Adams, A.M., Johnson, J.M., Tenge, B.J., Dang, N.L. and Wekell, M.M. 1999. An oligonucleotide-ligation assay for the differentiation between Cyclospora and Eimeria spp. polymerase chain reaction amplification products. J. Food Prot. 62(6), 682–685. Jordan, A.B., Blake, D., Beard, J., Beharry, A., Serrette, L., Soleyn, A., Sookhoo, J., Blake, L., Brown, G. and Oura, C. 2018. Molecular identification of Eimeria species in broiler chickens in Trinidad, West Indies. Vet. Sci. 5(1), 1–7. Kanyari, P. W.N., Kagira, J.M. and Mhoma, R.J. 2009. Prevalence and intensity of endoparasites in small ruminants kept by farmers in Kisumu Municipality, Kenya. Livestock Res. Rural Dev. 21(11), 202. Kawahara, F., Zhang, G., Mingala, C.N., Tamura, Y., Koiwa, M., Onuma, M. and Nunoya, T. 2010. Genetic analysis and development of species-specific PCR assays based on ITS-1 region of rRNA in bovine Eimeria parasites. Vet. Parasitol. 174(1–2), 49–57. Khodakaram-Tafti, A., Hashemnia, M., Razavi, S.M., Sharifiyazdi, H. and Nazifi, S. 2013. Genetic characterization and phylogenetic analysis of Eimeria arloingi in Iranian native kids. Parasitol. Res. 112(9), 3187–3192. Lassen, B. and Järvis, T. 2009. Eimeria and Cryptosporidium in Lithuanian cattle farms. Vet. Med. Zoot. 48(70), 24–28. Liang, G., Yang, X., Liu, D., Li, Y., Wang, J., Chen, X., Zhao, G. and Song, J. 2022. Molecular characterization of 18S rRNA, ITS-1, ITS-2, and COI from Eimeria christenseni and E. arloingi in goats from Shaanxi Province, Northwestern China. Animals 12(11), 1340 . López-Osorio, S., Chaparro-Gutiérrez, J.J. and Gómez-Osorio, L.M. 2020. Overview of poultry Eimeria life cycle and host-parasite interactions. Front. Vet. Sci. 7, 1–8. Martin, S.W., Meek A.H. and Willeberg, P. 1987. Veterinary epidemiology principles and methods. Ames, IA: Iowa States University Press. pp: 23–40. Matsubayashi, M., Takami, K., Abe, N., Kimata, I., Tani, H., Sasai, K. and Baba, E. 2005. Molecular characterization of crane Coccidia, Eimeria gruis and E. reichenowi, found in feces of migratory cranes. Parasitol. Res. 97(1), 80–83. Mohamaden, W.I., Sallam, N.H. and Abouelhassan, E.M. 2018. Prevalence of Eimeria species among sheep and goats in Suez Governorate, Egypt. Int. J. Vet. Sci. Res. 6(1), 65–72. Nguyen-Ho-Bao, T., Lu, T.A. and Huu Nguyen, H. 2022. Studies on molecular characteristics of Eimeria arloingi in goats (Capra hircus) in Vietnam. Int. J. Eng. Sci. Technol. 10(1), 19–28. Pamungkas, P.A., Apsari, I.A.P. and Widyastuti, S.K. 2021. Prevalensi Infeksi Eimeria spp. yang Tinggi pada Kambing yang Dipelihara di Kota Denpasar. Indonesia. Med. Veterinus. 10(6), 861–868. Pertiwi, V.R., Riffiandi, N. and Sofiana, A. 2023. Infeksi Parasit Gastrointestinal pada Kambing (Capra aegagrus hircus) di Desa Rajabasa Lama Kabupaten Lampung Timur. PETERPAN 5(1), 10–15. Pyziel, A.M., Demiaszkiewicz, A.W., Osińska, B., Dolka, I., Anusz, K. and Laskowski, Z. 2020. Usefulness of PCR–RFLP of 18S rRNA gene for rapid post-mortem diagnostics of highly pathogenic Eimeria spp. (Apicomplexa: Eimeriidae) of European bison, Bison bonasus L. with histopathological correlation. Int. J. Parasitol. Parasites. Wildl. 12, 13–18. Rehman, T.U., Khan, M.N., Sajid, M.S., Abbas, R.Z., Arshad, M., Iqbal, Z. and Iqbal, A. 2010. Epidemiology of Eimeria and associated risk factors in cattle of district Toba Tek Singh, Pakistan. Parasitol. Res. 108(5), 1171–1177. Roh, S.G., Kim, J., Ku, B.K. and Lee, K. 2023. Case study: pathological and phylogenetic analysis of coccidiosis in two goats with heavy infection of unrecorded Eimeria sp. Parasitol. Int. 92, 102662. Ruiz, A., Guedes, A.C., Muñoz, M.C., Molina, J.M., Hermosilla, C., Martín, S., Hernández, Y.I., Hernández, Á., Pérez, D., Matos, L., López, A.M. and Taubert, A. 2012. Control strategies using diclazuril against coccidiosis in goat kids. Parasitol. Res. 110(6), 2131–2136. Souza, L.E.B., da Cruz, J.F., Teixeira Neto, M.R., Albuquerque, G.R., Melo, A.D.B. and Tapia, D.M.T. 2015. Epidemiology of Eimeria infections in sheep raised extensively in a semiarid region of Brazil. Rev. Bras. Parasitol. Vet. 24(4), 410–415. Stotish, R.L., Wang, C.C. and Meyenhofer, M. 1978. Structure and composition of the oocyst wall of Eimeria tenella. J. Parasitol. 64(6), 1074–1081. Takeo, T., Tanaka, T., Matsubayashi, M., Maeda, H., Kusakisako, K., Matsui, T., Mochizuki, M. and Matsuo, T. 2014. Molecular and phylogenetic characterizations of an Eimeria krijgsmanni Yakimoff and Gouseff, 1938 (Apicomplexa: Eimeriidae) mouse intestinal protozoan parasite by partial 18S ribosomal RNA gene sequence analysis. Parasitol. Int. 63(4), 627–630. Tenrisanna, V. and Kasim, K. 2021. Livestock farming income analysis of farm households in Indonesia. IOP Conf. Ser. Earth Environ. Sci. 788(1), 1–6 . Underwood, W.J., Blauwiekel, R., Delano, M.L., Gillesby, R., Mischler, S.A. and Schoell, A. 2015. Chapter 15—Biology and diseases of ruminants (sheep, goats, and cattle). In Laboratory animal medicine, 3rd ed. Eds. Anderson, L.C., Fox, J.G., Otto, G., Pritchett-Corning, K.R. and Whary, M.T. Cambridge, MA: Academic Press, pp: 623–694. Verma, R., Sharma, D.K., Gururaj, K., Paul, S., Banerjee, P.S. and Tiwari, J. 2017. Molecular epidemiology and point mutations in ITS1 and 18S rRNA genes of Eimeria ninakohlyakimovae and E. christenseni isolated from Indian goats. Vet. Parasitol. Reg. Stud. 9, 51–62. | ||
| How to Cite this Article |
| Pubmed Style Dewi DA, Nugraheni YR, Awaludin A, Ninditya VI, Priyowidodo D, Nurcahyo RW, Ekawasti F, Prastowo J. First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia. Open Vet. J.. 2025; 15(1): 139-150. doi:10.5455/OVJ.2025.v15.i1.13 Web Style Dewi DA, Nugraheni YR, Awaludin A, Ninditya VI, Priyowidodo D, Nurcahyo RW, Ekawasti F, Prastowo J. First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia. https://www.openveterinaryjournal.com/?mno=220662 [Access: September 04, 2025]. doi:10.5455/OVJ.2025.v15.i1.13 AMA (American Medical Association) Style Dewi DA, Nugraheni YR, Awaludin A, Ninditya VI, Priyowidodo D, Nurcahyo RW, Ekawasti F, Prastowo J. First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia. Open Vet. J.. 2025; 15(1): 139-150. doi:10.5455/OVJ.2025.v15.i1.13 Vancouver/ICMJE Style Dewi DA, Nugraheni YR, Awaludin A, Ninditya VI, Priyowidodo D, Nurcahyo RW, Ekawasti F, Prastowo J. First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia. Open Vet. J.. (2025), [cited September 04, 2025]; 15(1): 139-150. doi:10.5455/OVJ.2025.v15.i1.13 Harvard Style Dewi, D. A., Nugraheni, . Y. R., Awaludin, . A., Ninditya, . V. I., Priyowidodo, . D., Nurcahyo, . R. W., Ekawasti, . F. & Prastowo, . J. (2025) First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia. Open Vet. J., 15 (1), 139-150. doi:10.5455/OVJ.2025.v15.i1.13 Turabian Style Dewi, Dias Aprita, Yudhi Ratna Nugraheni, Aan Awaludin, Vika Ichsania Ninditya, Dwi Priyowidodo, Raden Wisnu Nurcahyo, Fitrine Ekawasti, and Joko Prastowo. 2025. First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia. Open Veterinary Journal, 15 (1), 139-150. doi:10.5455/OVJ.2025.v15.i1.13 Chicago Style Dewi, Dias Aprita, Yudhi Ratna Nugraheni, Aan Awaludin, Vika Ichsania Ninditya, Dwi Priyowidodo, Raden Wisnu Nurcahyo, Fitrine Ekawasti, and Joko Prastowo. "First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia." Open Veterinary Journal 15 (2025), 139-150. doi:10.5455/OVJ.2025.v15.i1.13 MLA (The Modern Language Association) Style Dewi, Dias Aprita, Yudhi Ratna Nugraheni, Aan Awaludin, Vika Ichsania Ninditya, Dwi Priyowidodo, Raden Wisnu Nurcahyo, Fitrine Ekawasti, and Joko Prastowo. "First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia." Open Veterinary Journal 15.1 (2025), 139-150. Print. doi:10.5455/OVJ.2025.v15.i1.13 APA (American Psychological Association) Style Dewi, D. A., Nugraheni, . Y. R., Awaludin, . A., Ninditya, . V. I., Priyowidodo, . D., Nurcahyo, . R. W., Ekawasti, . F. & Prastowo, . J. (2025) First molecular detection of Eimeria spp. in domestic goats from Java Island, Indonesia. Open Veterinary Journal, 15 (1), 139-150. doi:10.5455/OVJ.2025.v15.i1.13 |