| Original Article | ||
Open Vet J. 2022; 12(2): 273-280 Open Veterinary Journal, (2019), Vol. 12(2): 273–280 Original Research Molecular detection and characterization of Orf virus from goats in EgyptAyman Ahmed Shehata1*, Hussein Abdalatif Elsheikh2 and Eman Beshry Abd-Elfatah11Department of Animal Medicine, Infectious Diseases, Faculty of Veterinary Medicine, Zagazig University, El-Shohada, Moawwad, Qesm Awel AZ Zagazig, 44519, Egypt 2The Veterinary Clinic, Faculty of Veterinary Medicine, Zagazig University, El-Shohada, Moawwad, Qesm Awel AZ Zagazig, 44519, Egypt *Corresponding Author: Ayman Ahmed Shehata. Department of Animal Medicine, Infectious Diseases, Faculty of Veterinary Medicine, Zagazig University, El-Shohada, Moawwad, Qesm Awel AZ Zagazig, 44519, Egypt. Email: aymanshehata305 [at] yahoo.com Submitted: 05/01/2022 Accepted: 26/03/2022 Published: XX/04/2022 © 2022 Open Veterinary Journal
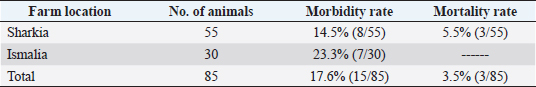
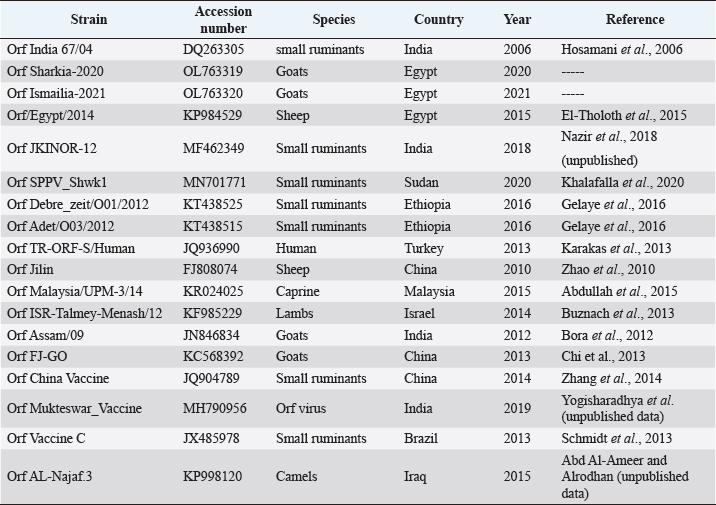
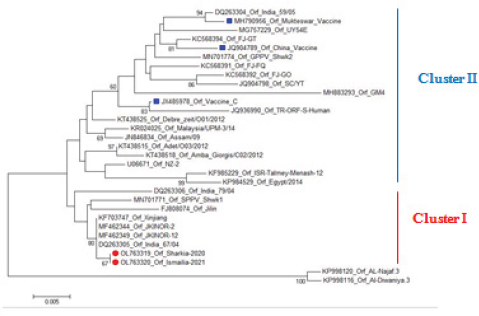
AbstractBackground: Orf is a highly contagious viral skin disease in sheep and goats caused by Orf virus (ORFV) in the genus Parapoxvirus. Although sheep and goats are considered an essential food resource, particularly in Africa, ORFV infection represents an increasing challenge to animal productivity causing high economic losses. Aim: This study aimed to detect and characterize the ORFV in suspected clinically diseased goats in two neighboring Egyptian governorates, Al-Sharkia and Ismailia, flocks during April 2020 and July 2021by using PCR and phylogenetic analysis of partial B2L sequence. he present study indicate the necessity for establishing normal heart values in conscious and anaesthetized individuals. Methods: Kids from two Egyptian governorates showed the clinical picture of ORFV infection. Samples were collected (n=15) from two different flocks during April 2020 and July 2021. PCR was carried out to detect the ORFV by targeting a highly conserved sequence within ORFV (B2L) gene. To determine the phylogenetic relationship with other ORFV strains, sequencing and phylogenetic analysis were performed. Results: ORFV infection was confirmed in 12 samples of oral scabs (80%) by PCR targeting a highly conserved sequence within B2L gene. Sequencing of DNA products was performed and obtained sequences revealed 100% identity at the nucleotide level. Two ORFVs, one from each outbreak showed 98.2% nucleotide identity with a previous Egyptian ORFV (KP984529) whereas our isolates showed higher nucleotide identities, 99.1% and 98.7% with ORFV strains from neighboring countries, Sudan and Ethiopia, respectively. The phylogenetic tree grouped isolates into two main clusters, cluster I included isolates of this study and foreign ones mainly from China, India, and Sudan. Interestingly, the vaccine strains of ORF used in different countries were grouped in cluster II with previous Egyptian isolate (KP984529), Ethiopian and Israeli ORFV isolates. Conclusion: Molecular characterization of B2L gene of ORFV isolates revealed higher sequence identities and more close genetic relationships with other ORFV strains circulating in neighboring countries than with the Egyptian isolates. These findings provide an insight into the genetic diversity of field ORFV isolates circulating in goats in the Egyptian governorates. Keywords: Egypt, Orf virus, Goat, B2L gene, PCR. IntroductionAlthough sheep and goats represent an essential food resource for millions of people, particularly in Africa, increasing challenges, including infectious diseases are confronting animal productivity. In sheep and goats, parapoxviruses (PPVs) infection is circulated worldwide and is known as sore mouth, ORF, Contagious Pustular Dermatitis (CPD), Contagious Ecthyma (CE), causing enormous economic loss (Khalafalla et al., 2020). The disease is caused by Orf virus (ORFV), a linear dsDNA, a prototype member in the genus Parapoxvirus under the subfamily Chordopoxvirinae of the Poxviridae family (Haig and Mercer, 1998). The viral genome is generally organized into conserved central portion and variable terminal regions, which encode the molecules required for viral interactions with host cells (Fleming et al., 2015). The central conserved portion involves several genes that are responsible for virus replication including the B2L gene which encodes for a major immunogenic envelope protein that is a homologue of vaccinia virus (Gelaye et al., 2016; Olivero et al., 2018). Orf is a highly contagious viral skin disease of farmed small ruminants, camels, several wild ruminants and humans (McElroy and Bassett, 2007; Ali et al., 2013), and the disease usually hits lambs and kids aged from 3 to 6 months but neonatal lambs and kids around the age of two weeks show severe clinical manifestations (Oem et al., 2009). Clinically, ORFV mainly infects epithelial cells and is associated with cutaneous lesions manifested through different stages as maculopapular, vesicular pustules and scabby proliferative lesions that mainly affect the skin around the lips, oral and nasal mucosa and udders (Adedeji et al., 2018). The economic losses associated with virus outbreaks are cost-effective as a result of growth delay, reduction of the animal’s body condition and the secondary bacterial or fungal infections that can complicate cases and reach the mortalities up to 93% (Zhao et al., 2010; Gelaye et al., 2016; Bala et al., 2019). In Egypt, the disease was previously reported in goats and sheep from different localities by using different conventional diagnostic tools. In 2010, sheep from different farms in Ismailia governorate developed warts like lesions on the lips, gum and teats, where ORFV was isolated and confirmed in nine samples of scabs (60%) using the tissue culture in lamb kidney cells, corioallantoic membrane (CAM) of emberyonated chicken egg and agar gel preciptation test (Said et al., 2013). Real time PCR was found to be more sensitive than virus isolation and conventional PCR in detecting Orf infection in a herd of native breed sheep in Qalubia province where ORFV was found in 100%, 80% and 70% of samples by real time PCR, virus isolation and conventional PCR, respectively (Selim et al., 2016). In the Minufiya governorate, ORFV was detected in grassing sheep and goats suffering from scabs and nodules at oral commissures and gums with PCR targeting the late transcription factor gene (VLTF-1) in 7 out of 10 samples (70%) by Khalil et al. (2021). Laboratory diagnosis of ORFV has conventionally based on observing the characteristic clinical signs and further confirmation by virus isolation, histopathology and electron microscopical examination of negatively stained scabs with the characteristic ovoid-shape of the virion. Now, PCR is widely used as a specific and sensitive method for virus detection worldwide (Chan et al., 2007). Regarding PCR targeting genes, some studies depended on VLTF-1 gene to detect and characterize ORFV in Egypt, as in the study performed by Mahmoud et al. (2010) who successfully amplified VLTF-1 gene from 77.5% of sheep and goats skin samples. Also, the p55 gene (H3L genomic region) was used to confirm ORFV from four Egyptian governorates in skin samples following the virus isolation on embryonated chicken eggs and MDBK cell culture. Notably, B2L gene has been used extensively in molecular detection and diagnosis as well as phylogenic analyses of various ORFV isolates worldwide including Egypt (Zhang et al., 2010; El-Tholoth et al., 2015; Selim et al., 2016). Therefore, the aim of this study was to detect and characterize ORFV in suspected clinically diseased goats from Al-Sharkia and Ismailia governorates during April 2020 and July 2021by using PCR and phylogenetic analysis of partial sequence of the B2L gene. Materials and MethodsAnimals (Field study)Goats from two small local private farms were included in this study. One farm was located in Dahmasha, Belbis, Sharkia governorate, in the north part of Egypt, with 55 native goats (34 adults and 21 kids aged from two weeks to four months). During a field visit in April 2020 to the herd, a total of eight kids were found suffering from suspected Orf signs. Also, the collected herd data revealed a history of three dead cases (5.5%). In July 2021, seven kids aged two weeks to about two months showed proliferative scabby lesions at the oral commissures with mild fever in a flock consisting of 30 goats with a wide age structure from less than one month to adults located in El-Tell El Kebir, Ismailia Governorate, Egypt. Notably, other animals in both flocks of different age groups did not show any clinical signs. Owners of each flock were advised to quickly isolate infected kids to prevent the spread of infection to other animals. Diseased kids were symptomatically treated with a broad spectrum antibiotic (oxytetracycline 10% at 10mg /kg B.Wt) for three days to prevent complications with secondary bacterial infections. Also, oral lesions were sprayed with gentian violet and supportive treatment with intramuscular multivitamins and intravenous dextrose saline fluid therapy was applied in cases that were unable to suckle their dams. Sample collectionField visits were conducted to collect samples as well as the data of the two herds. In both farms, sample collection was restricted to the cases showing a suspected clinical picture of ORFV infection for further confirmatory laboratory diagnosis. As a result, a total of 15 oral necrotic tissues and scab samples were collected from the two farms. The samples were taken in a sterile dry tube and triturated in PBS pH 7.4 with antibiotics and antifungal to make up a 10 % (w/v) solution. Then, samples were ground by using TissueLyser LT (Qiagen, Hilden, Germany) and were centrifuged for 10 min at 4oC. The supernatant was collected for PCR detection and identification of ORFV. Extraction of DNATotal DNA was extracted from collected clinical samples by using the QIAamp DNA kit (QIAGEN, Germany) following the manufacturer’s instructions. Extracted DNA was stored at - 20 çC until being used. Polymerase chain reaction (PCR)PCR was carried out to detect ORFV by using Emerald Amp PCR Master Mix (Takara, Japan), in samples from suspected diseased animals by targeting a highly conserved sequence within ORFV (B2L) gene using forward primer (PPP-1), 5-GTC GTC CAC GAT GAG CAG CT-3 and reverse primer (PPP-4),5-TAC GTG GGA AGC GCC TCG CT-3, the primer was designed to amplify a specific segment of 594 bp (Inoshima et al., 2000). Primers were synthetized by Metabion International AG Company. Cycling conditions were as follows: the reaction was applied in a 25 μl reaction volume containing 4.5 μl of PCR grade water, 6 μl of DNA template, 1 μM of each primer, and 12.5 μl of Emerald Amp PCR master mix. Primary denaturation was performed at 94 0C for 5 min, secondary denaturation at 94 °C for 40 sec, annealing at 55 °C for 40sec, extension at 72 °C for 45 sec for 35 cycles and a final extension at 72 °C for 10 min. The amplified DNA products were resolved by 1.5% agarose gel electrophoresis, and were visualized by using a UV Transilluminator. Positive and/or negative controls were represented by field samples that were previously confirmed by PCR for the related gene at the reference laboratory, Animal Health Research Institute, Dokki, Egypt. Sequencing and phylogenetic analysisAmplified PCR products were purified according to the purification protocol of QIAquick PCR Product extraction kit. Sequencing in both directions was applied by using Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer) in Applied Biosystems3130 genetic analyzer (HITACHI, Japan). PCR products of two positive samples that had the higher band intensity, one positive sample from each farm, were used for sequencing. Once the sequence results were obtained, the sequences were subjected to NCBI-BLAST analysis (Basic Local Alignment Search Tool) (Altschul et al., 1990) to establish sequence identity to GenBank accessions. The phylogenetic analysis was performed by the MegAlign module of Lasergene DNA Star version 12.1 (Thompson et al., 1994) and phylogenetic tree was created by using maximum likelihood, neighbor joining method in MEGA program version 6.0 software (Tamura et al., 2013). Ethical approvalNo ethical approval was needed and collection of samples from clinically diseased animals was for the routine diagnosis of the disease under the usual veterinary service work in Egypt and according to the national standards within the country. ResultsClinical findingsIn the two farms, infection was restricted to young kids from less than one month to two months, while older animals were clinically normal at field examination. Morbidity and mortality rates were 14.5% and 23.35% in the first and second farms in Al-Sharkia and Ismailia governorates, respectively, whereas history from first farm revealed the mortality of three cases (5.5%) as shown in Table 1. Clinical signs of ORFV infection in goats varied from nodular lesions evolving in proliferative scabs formation developed around oral commissures and nostrils to ulcer and necrotic lesions in the inner side of upper and lower lips. In addition, salivation, emaciation and mild pyrexia were recorded and the oral lesions hindered animals to suckle and feed (Fig. 1). Diseased animals showed complete recovery after 10-15 days following the treatment. Virus detection in clinical samples by PCRPCR successfully detected ORFV in a total of 12 samples of oral lesions (80%) from the two herds, positive amplification of the ORFV B2L gene on agarose gel was photographed at 594 bp compared with positive control. Out of eight samples collected from the first herd, six samples (75%) were found positive, while one sample was negative for B2L gene in the second, with six samples showed virus positivity (85.7%). This study confirmed ORFV circulation in small ruminant flocks within the Egyptian governorates. Nucleotide sequencing and phylogenetic analysisAfter virus detection by PCR, sequencing of DNA products was performed for two representative samples, one from each different farm, representing isolates of ORFV in the two governorates. Furthermore, the two sequences were subjected to NCBI Blast analysis and confirmed as ORFV and then were deposited in GenBank under accession numbers OL763319 and OL763320. The two sequences shared 100% identity at the level of nucleotide sequences. The phylogenetic relationship of the two ORFVs with other ORFV strains was determined, and the deposited B2L gene sequences from GenBank that were used in this study are presented in Table 2. Results of comparing our sequences with other sequences from neighboring countries (Sudan, Ethiopia, and Israel), Asian and European countries (Turkey, Malaysia, Taiwan, China, and India) revealed no significant differences among the different ORF viruses (ORFVs) circulated in different parts of the world based on the viral B2L gene. The phylogenetic tree (Fig. 2) showed that the two isolates of this study were grouped together and were genetically closely related to Indian sheep ORFVs (MF462349, MF462344 and DQ263305) and virus isolated from goats in China (KF703747) with high percentage of identity (99.8%) with all these virus strains. Also, our Egyptian ORFVs were grouped in the same cluster with viruses from neighboring countries, MN701771 strain previously detected in Sudan with 99.1% identity. Goat isolates of this study showed the same percentage of identity (98.2%) with previous Egyptian ORFV (KP984529) identified from sheep in El-Beheira governorate, and with Israel ORFV (KF985229) reported by Buznach et al. (2013). Notably, strains of ORF vaccines, JQ904789, MH790956 and JX485978, from different countries were grouped in another cluster and shared 97.1%, 97.1% and 98.2% of identities with the two strains of the present study. Table 1. Epidemiological feature of suspected ORFV infections.
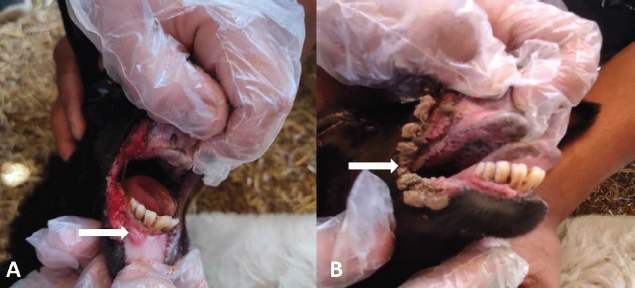
Fig. 1. Ulcer formation in inner side of lips (A). Proliferative scab lesions around oral commissures (B). Table 2. The phylogenetic relationship of our isolates with other ORFV deposited sequences from GenBank.
DiscussionOrf is an acute, zoonotic debilitating viral skin disease that affects domesticated and wild ruminants. In Egypt, although the disease has significant economic importance due to its contagious nature and high fatality in lambs and kids, especially on rural areas, the number of studies that have addressed in this topic is not in line with the rate of disease occurrence and spread. Moreover, there is no effective vaccine to control the disease in Egypt; therefore early detection and identification of the ORFV is necessary for effective control of the infection (Mahmoud et al., 2010).
Fig. 2. Phylogenetic tree constructed based on partial sequences of the B2L gene (594 bp) showing the genetic relationships between Egyptian ORFVs and other virus sequences from GenBank. The sequences of this study are marked with round red spot and vaccine isolates are marked with blue square. In this study, two farms from Al-Sharkia and Ismailia governorates showed the suspected clinical picture of ORFV infection in goats. In these outbreaks, morbidity rates were found to be 14.5% and 23.35% in the first and the second farms, respectively, while the mortality rate was 5.5% from the first farm. A similar morbidity rate of 15% and a case fatality rate of 4.7 % in investigated goats were reported in Sudan (Khalafalla et al., 2020). However, some previous studies revealed a higher morbidity rate of up to 40% in goats (Sevik, 2017), whereas a lower ORFV infection rate (4.8 %) was reported in Egypt in a flock of sheep and goats. These differences may be attributable to size of sample and sampling location. Also, higher mortality rates have been reported up to 90%, depending on host immunity, secondary complicated bacterial or viral infections and hygiene (Zhao et al., 2010; Gelaye et al., 2016; Bala et al., 2018). Although adult goats were found infected with Orf in previous studies (Khalafalla et al., 2020), our clinical examination in this study revealed that the disease hit mainly young kids aged less than two months as reported in previous studies (El-Tholoth et al., 2015; Sevik, 2017; Bala et al., 2018). Clinically, infected kids in both farms suffered from muco-cutaneous scabs around the lips, mouth, nostrils, the upper and lower eyelids and oral mucosa, necrosis and ulceration within the buccal cavity, which is reported in other studies (Maan et al., 2014; El-Tholoth et al., 2015; Srinivasa Babu et al., 2018; Lawal et al., 2021). Diseased animals showed complete recovery after 10-15 days following the treatment, as previously reported by Maan et al. (2014) and El-Tholoth et al. (2015). Although ORFV infection is suspected tentatively based on the characteristic clinical signs that can be used as a good reference due to an epitheliotropic nature of ORFV (Chan et al., 2007), the disease may be confused with other viral infections causing oral lesions (Watson, 2004). Therefore, PCR was used in this study to detect and further characterize virus field isolates based on ORFV011, which plays a role in the virus pathogenesis and encodes a major and highly immunogenic envelope protein (B2L) (Sullivan et al., 1994; Inoshima et al., 2001; Sevik, 2017). ORFV was identified in 12 samples of oral scabs out of 15 samples were examined. These results are in line with Maan et al. (2014), El-Tholoth et al. (2015) and Sowmiya et al. (2018). In this study, sequencing analysis of the B2L partial sequences revealed a 100% level of the nucleotide identity between the two viral sequences. In contrast, the similarity with Parapoxvirus isolates of sheep and goats from different geographic parts ranged from 95.8 to 99.8%. These results agree with Mahmoud et al. (2010), who used VTLF-1 gene to characterize ORFV from scabs of goats and sheep in the Giza governorate and alignment results revealed 100% identical virus isolates; these similarities between viral isolates detected at the same study level may be attributed to uncontrolled movement of animals between governorates. On the other hand, ORFVs from Ismailia, Menoufia and Sharkia governorates exposed lower homology percentage (92.8% - 95.3% ) based on P55 gene, where isolate Ismailia-1 possessed the higher variability compared with other two Egyptian isolates (Ali et al., 2013). While, a previous study showed a high identity between Egyptian (KP984529) and Israeli (KF985229) ORFV strains in 2015 (El-Tholoth et al., 2015), ORFV strains of this study were grouped in cluster I away from these two isolates with 98.2% nucleotide identity and instead were closely related to a Sudanese strain (MN701771) previously diagnosed by (Khalafalla et al., 2020) with 99.1% nucleotide identity. These findings may be associated with the fact that Egypt imports small ruminants from various African countries including Sudan and Ethiopia to overcome the deficiency in meat production in addition to uncontrolled animal movement within governorates, hence; it is more likely that ORFV sequences circulating in Egypt appears to be from different origins. Regarding the B2L gene-based phylogenetic tree, ORFV sequences are grouped into two main clusters. While, Cluster I included Egyptian isolates of this study and foreign isolates mainly from China, India, and neighboring countries as Sudan, vaccinal ORF strains, JQ904789, MH790956 and JX485978, used in different countries such as India, China and Brazil were grouped in cluster II. These findings suggest that the high degree of similarity of the two sequences with vaccine strains at the nucleotide sequence level might be because of sequencing of highly conserved genes rather than a close relationship between field and vaccine viruses. Taken together with the previous finding that the co-circulation of vaccinal strains and field ones is very unlikely in the same geographic areas, the vaccination strategies against the disease can be highly implemented. This study focused on the molecular detection and characterization of ORFV infection to figure out the viral sequences circulating in Al-Sharkia and Ismailia governorates in two outbreaks during April 2020 and July 2021; however, other diagnostic techniques of viral isolation are highly required to identify virus morphology and understand the relationship to the other ORFV isolates circulating within the country. ConclusionOrf disease was confirmed in goats from Al-Sharkia and Ismailia governorates showing the suspected clinical picture of ORFV infection. In these outbreaks, the overall morbidity rate was found to be 17.6%, while the mortality rate was 5.5%. Molecular detection and further sequence analysis of B2L gene in ORFV isolates revealed a close genetic relationship with other ORFV strains circulating worldwide with particular reference to Indian, Chinese and Sudanese isolates. While isolates of this study showed 98.2% nucleotide identity with a previous Egyptian ORFV (KP984529), higher nucleotide identities 99.1% and 98.7% were detected with ORFV strains from neighboring countries, Sudan and Ethiopia, respectively. Two main clusters were found in the phylogenetic tree, Cluster I included isolates of this study and foreign isolates mainly from China, India, and Sudan. The vaccine ORFVs used in different countries were grouped in cluster II with previous Egyptian isolate (KP984529), Ethiopian and Israeli ORFV isolates. This study was based on B2L gene however; further studies are required to study the virus at the level of more genes to improve tracking and understanding of ORFV strains circulation in Egypt. Conflict of interestThe authors declare that they have no competing interests. Authors’ contributionsAAS and HAE: Collected the samples. AAS, HAE and EBA processed all the laboratory work and drafted and revised the manuscript. All authors read and approved the final manuscript. ReferencesAbdullah, A.A., Ismail, M.F., Balakrishnan, K.N., Bala, J.A., Hani, H., Abba, Y., Awang Isa, M.K., Abdullah, F.F., Arshad, S.S., Nazariah, Z.A., Abdullah, R., Mustapha, N.M. and Mohd-Lila, M.A. 2015. Isolation and phylogenetic analysis of caprine Orf virus in Malaysia. Virus Dis. 26(4), 255-259. Adedeji, A.J., Adole, J.A. and Chima, N.C. 2018. Contagious ecthyma in three flocks of goats in Jos-south LGA, Plateau State, Nigeria. Sokoto J.Vet. Sci. 16(1), 107-112. Ali, M.H., Ahmed, M.H., Tamam, S.M., Arafa, A., Saad, A. A., Ali, W.F. and Madbouly, H.M. 2013. Molecular Characterization of Orf Virus Isolated from Sheep and Goats in Egypt. Global Vet. 11(1), 98-106. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipmanl, D.J. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403-410. Bala, A.J., BalaKrishnan, N.K. and Firdaus, F.J. 2019. Identification of strain diversity and phylogenetic analysis based on two major essential proteins of orf viruses isolated from several clinical cases reported in Malaysia. Infect. Genet. Evol. 77, 104076. Bala, J.A., Balakrishnan, K.N., Abdullah, A.A., Mohamed, R., Haron, A.W., Jesse, F.F.A., Noordin, M.M. and Mohd-Azmi, M.L. 2018. The re-emerging of orf virus infection: a call for surveillance, vaccination and effective control measures. Microb. Pathog. 120, 55–63. Bora, D.P., Barman, N.N., Das, S.K., Bhanuprakash, V., Yogisharadhya, R., Venkatesan, G., Kumar, A., Rajbongshi, G., Khatoon, E., Chakraborty, A. and Bujarbaruah, K.M. 2012. Identification and phylogenetic analysis of orf viruses isolated from outbreaks in goats of Assam, a northeastern state of India. Virus Genes 45(1), 98-104. Buznach, A., Hahn, S., Stram, Y., Menasherov, S., Edery, N., Shicaht, N., Kenigswald, G. and Perl, S. 2013. A Case Report: Contagious Ecthyma - deviations in the Anatomical Appearance of Lesions in an Outbreak in Lambs in Israel. Israel J. Vet. Med. 68(4), 246-251. Chan, K.W., Lin, J.W., Lee, S.H., Liao, C.J., Tsai, M.C., Hsu, W.L., Wong, M.L. and Shih, H.C. 2007. Identification and phylogenetic analysis of orf virus from goats in Taiwan. Virus Genes 35, 705–712. Chi, X., Zeng, X., Hao, W., Li, M., Li, W., Huang, X., Wang, S. and Luo, S. 2013. Heterogeneity among Orf Virus Isolates from Goats in Fujian Province, Southern China. PLoS One 8(10), E66958. El-Tholoth, M., Elnaker, Y.F. and Shiha, G. 2015. Phylogenetic analysis of B2L gene of Egyptian orf virus from naturally infected sheep. Virus Dis. 26(3), 147–150. Fleming, S.B., Wise, L.M. and Mercer, A.A. 2015. Molecular genetic analysis of orf virus: a poxvirus that has adapted to skin. Viruses 7, 1505–1539. Gelaye, E., Achenbach, J.E., Jenberie, S., Ayelet, G. and Belay, A. 2016. Molecular characterization of orf virus from sheep and goats in Ethiopia, 2008–2013. Virol. J. 13, 34. https://doi.org/10.1186/s12985-016-0489-3. Haig, D.M. and Mercer, A. A. 1998. Ovine diseases. Orf. Vet. Res. 29, 311–326. Hosamani, M., Bhanuprakash, V., Scagliarini, A. and Singh, R.K. 2006. Comparative sequence analysis of major envelope protein gene (B2L) of Indian orf viruses isolated from sheep and goats. Vet. Microbiol. 116(4), 317-324. Inoshima, Y., Morooka, A. and Sentsui, H. 2000. Detection and diagnosis of parapoxvirus by the polymerase chain reaction. J. Virol. Methods 84(2), 201-208. Inoshima, Y., Murakami, K., Yokoyama, T. and Sentsui, H. 2001. Genetic heterogeneity among parapoxviruses isolated from sheep, cattle and Japanese serows (Capricornis crispus). J. Gen. Virol. 82, 1215-1220. Karakas, A., Oguzoglu, T.C., Coskun, O., Artuk, C., Mert, G., Gul, H.C., Sener, K. and Ozkul, A. 2013. First molecular characterization of a Turkish orf virus strain from a human based on a partial B2L sequence. Arch. Virol. 158(5), 1105-1108. Khalafalla, A.I., Elhag, A.E. and Ishag, H.Z.A. 2020. Field investigation and phylogenetic characterization of orf virus (ORFV) circulating in small ruminants and Pseudocowpoxvirus (PCPV) in dromedary camels of eastern Sudan. Heliyon 6(3), e03595. Khalil, A.E., Zeinab, R.A., Fouad, S.E., Ayman, S.E. and Saad, S.S. 2021. Molecular and serological diagnosis of Orf virus from sheep and goats in Minufiya governorate, Egypt. Benha Vet. Med. J. 41, 55-59. Lawal, N., Ibrahim, M., Onawala, D.A., Bello, M.B., Aliyu, R.M., Baraya, Y.S., Aliyu, A., Ibrahim, A.M. and Sa’adu, A. 2021. Molecular characterization and phylogenetic analysis of orf virus isolated from goats in Sokoto Metropolis, Nigeria. Future Sci. OA. FSO700, 2056-5623. https://doi.org/10.2144/fsoa-2020-0162. Maan, S., Kumar, A., Batra, K., Singh, M., Nanda, T., Ghosh, A. and Maan, N.S. 2014. Isolation and molecular characterization of contagious pustular dermatitis virus from Rajasthan, India. Virus Dis. 25(3), 376–380. Mahmoud, M., Abdelrahman, K. and Soliman H. 2010. Molecular and virological studies on contagious pustular dermatitis isolates from Egyptian sheep and goats. Res. Vet. Sci. 89, 290–294. McElroy, M.C. and Bassett, H.F. 2007. The development of oral lesions in lambs naturally infected with orf virus. Vet. J. 174, 663-664. Oem, J.K., Roh, I.S., Lee, K.H., Lee, K.K., Kim, H.R. and Jean, Y.H. 2009. Phylogenetic analysis and characterization of Korean orf virus from dairy goats: case report. Virol. J. 6, 167. Olivero, N., Reolon, E., Arbiza, J. and Berois, M. 2018. Genetic diversity of orf virus isolated from sheep in Uruguay. Arch. Virol. 163(5), 1285-1291. Said, A., Mohamed, S.I., AbdElhamid, N.K., Hosny, W.A. and Baheeg, E.M. 2013. Trials for Isolation of Contagious Pustular Dermatitis Virus (CPDV) from Sheep in Ismailia Governorate. Res. Zool. 3(1), 10-14. Schmidt, C., Cargnelutti, J.F., Brum, M.C., Traesel, C.K., Weiblen, R. and Flores, E.F. 2013. Partial sequence analysis of B2L gene of Brazilian orf viruses from sheep and goats. Vet. Microbiol. 162(1), 245-253. Selim, A., Elhaig, M., Höche, J. and Gaede, W. 2016. Molecular detection and analysis of Sheeppox and Orf viruses isolated from sheep from Qalubia, Egypt. Berl. Munch. Tierarztl. Wochenschr. 129(7-8), 310-317. Sevik, M. 2017. Association of two clusters of Orf virus isolates in outbreaks of infection in goat in the Central Anatolian region of Turkey. Virus Dis. 28(3), 345–348. Sowmiya, P.V., Ramya, K., Sukmar, K. and Madheswaran, R. 2018. Molecular Confirmation of Contagious Ecthyma Among Goats in Dharmapuri District of Tamil Nadu, India. Int. J. Curr. Microbiol. App. Sci. 7(6), 60-65. Srinivasa Babu, T., Rathnamma, D., Shrikrishna, I., Chandranaik, B.M., Veeregowda, B.M. and Manjunath Reddy, G.B. 2018. Diagnosis of orf virus infection in sheep and goats PCR and sequencing. J. Experi. Biol. Agri. Sci. 6(1), 176–187. Sullivan, J.T., Fleming, S.B. and Robinson A.J. 1994. Identification and characterization of an orf homologue of the vaccinia virus gene encoding the major envelope antigen p37K. Virol. J. 202, 968–973. Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. Thompson, J.D., Higgins, D.G. and Gibson, T.J. 1994. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucl. Aci. Res. 22, 4673-4680. Watson, P. 2004. Differential diagnosis of oral lesions and FMD in sheep. In Practice 26, 182–191. Zhang, K., Liu, Y., Kong, H., Shang, Y. and Liu, X. 2014. Comparison and phylogenetic analysis based on the B2L gene of orf virus from goats and sheep in China during 2009-2011. Arch. Virol. 159(6), 1475-1479. Zhang, K., Lu, Z. and Shang, Y. 2010. Diagnosis and phylogenetic analysis of orf virus from goats in China: a case report. Virol. J. 7(78), 1–5. Zhao, K., Song, D., He, W., Lu, H., Zhang, B., Li, C., Chen, K. and Gao, F. 2010. Identification and phylogenetic analysis of an Orf virus isolated from an outbreak in sheep in the Jilin province of China. Vet. Microbiol. 142, 408–415. | ||
| How to Cite this Article |
| Pubmed Style Shehata AA, Elsheikh HA, Abd-Elfatah EB, . Molecular detection and characterization of Orf virus from goats in Egypt. Open Vet J. 2022; 12(2): 273-280. doi:10.5455/OVJ.2022.v12.i2.16 Web Style Shehata AA, Elsheikh HA, Abd-Elfatah EB, . Molecular detection and characterization of Orf virus from goats in Egypt. https://www.openveterinaryjournal.com/?mno=140682 [Access: April 18, 2024]. doi:10.5455/OVJ.2022.v12.i2.16 AMA (American Medical Association) Style Shehata AA, Elsheikh HA, Abd-Elfatah EB, . Molecular detection and characterization of Orf virus from goats in Egypt. Open Vet J. 2022; 12(2): 273-280. doi:10.5455/OVJ.2022.v12.i2.16 Vancouver/ICMJE Style Shehata AA, Elsheikh HA, Abd-Elfatah EB, . Molecular detection and characterization of Orf virus from goats in Egypt. Open Vet J. (2022), [cited April 18, 2024]; 12(2): 273-280. doi:10.5455/OVJ.2022.v12.i2.16 Harvard Style Shehata, A. A., Elsheikh, H. A., Abd-Elfatah, E. B. & (2022) Molecular detection and characterization of Orf virus from goats in Egypt. Open Vet J, 12 (2), 273-280. doi:10.5455/OVJ.2022.v12.i2.16 Turabian Style Shehata, Ayman Ahmed, Hussein Abdalatif Elsheikh, Eman Beshry Abd-Elfatah, and . 2022. Molecular detection and characterization of Orf virus from goats in Egypt. Open Veterinary Journal, 12 (2), 273-280. doi:10.5455/OVJ.2022.v12.i2.16 Chicago Style Shehata, Ayman Ahmed, Hussein Abdalatif Elsheikh, Eman Beshry Abd-Elfatah, and . "Molecular detection and characterization of Orf virus from goats in Egypt." Open Veterinary Journal 12 (2022), 273-280. doi:10.5455/OVJ.2022.v12.i2.16 MLA (The Modern Language Association) Style Shehata, Ayman Ahmed, Hussein Abdalatif Elsheikh, Eman Beshry Abd-Elfatah, and . "Molecular detection and characterization of Orf virus from goats in Egypt." Open Veterinary Journal 12.2 (2022), 273-280. Print. doi:10.5455/OVJ.2022.v12.i2.16 APA (American Psychological Association) Style Shehata, A. A., Elsheikh, H. A., Abd-Elfatah, E. B. & (2022) Molecular detection and characterization of Orf virus from goats in Egypt. Open Veterinary Journal, 12 (2), 273-280. doi:10.5455/OVJ.2022.v12.i2.16 |