| Original Article | ||
Open Vet J. 2023; 13(4): 419-426 Open Veterinary Journal, (2023), Vol. 13(4): 419–426 Original Research Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmissionMaría Soledad Serena1,2, Javier Cappuccio2,3, Melisa Fossaroli4, Macarena Marta Williman1,5, Marina Dibarbora6, Renata Brizzio7,8, Germán Ernesto Metz1,2, Carolina Gabriela Aspitia1, Alejandro Perez9, Bruno Nicolás Carpinetti10 and María Gabriela Echeverría1,2*1Laboratorio de Virología, Facultad de Ciencias Veterinarias, Universidad Nacional de La Plata, La Plata, Argentina 2CONICET, CABA, Argentina 3Estación Experimental Agropecuaria Marcos Juárez, Marcos Juárez, Argentina 4Departamento de Patología, Facultad de Ciencias Veterinarias, Universidad Nacional de Rosario, Rosario, Argentina 5Becaria de la Agencia Nacional de Promoción Científica y Tecnológica, La Plata, Argentina 6Cátedra de Histología II y Embriología Especial, Facultad de Ciencias Veterinarias, Universidad Nacional de Rosario, Rosario, Argentina 7Patología General Veterinaria, Universidad Católica de Córdoba, Córdoba, Argentina 8Patología General Veterinaria, Universidad Nacional de Río Negro, Viedma, Argentina 9Programa Sanitario Porcino, Dirección Nacionad de Sanidad Animal, SENASA, CABA, Argentina 10Instituto de Ciencias Sociales y Administración, Universidad Nacional Arturo Jauretche, Florencio Varela, Argentina *Corresponding Author: María Gabriela Echeverría. Laboratorio de Virología, Facultad de Ciencias Veterinarias, Universidad Nacional de La Plata, La Plata, Argentina. Email: mariagabrielaecheverria [at] yahoo.com.ar Submitted: 24/10/2022 Accepted: 10/03/2023 Published: 07/04/2023 © 2023 Open Veterinary Journal
AbstractBackground: Aujeszky's disease is mainly a swine disease, still endemic worldwide. It can infect other mammalians, including human beings, and it is usually fatal with nervous symptoms. Ever since the disease was detected in 1988 in Argentina, many outbreaks have been reported involving both feral swine and dogs. Aim: At present, in Argentina, Pseudorabies virus (PRV) cases are sporadically reported; however, clinical cases are informed. This study aims to obtain information about the seroprevalence of PRV in wild boars and to isolate and characterize PRV from clinical samples. Methods: From 2018 to 2019, 78 wild boars’ serum samples from Bahía de Samborombón natural reserve were analyzed for antibodies to PRV using a virus neutralization test. Clinical samples from 17 pigs, 2 wild boars, 1 dog, and 1 cat were collected from 2013 to 2019 for viral isolation and detection of the presence of the gD gene by PCR. For sequence analysis, the gC partial gene was amplified. Results: Five strains were isolated from the dog, cat, and swine samples. The new PRV strains identified were confirmed by BLAST analysis, which revealed between 99.74% and 100% of similarity to the NIA-3 strain and phylogenetic analysis of the partial gene encoding the gC protein revealed that the PRV strains have divided into two main clades, clade 1 and clade 2. Conclusion: This report informed that most new cases of PRV were detected in the central regions of Argentina, where pig production is concentrated. The study in Bahía de Samborombón revealed a high percentage of detection but, the sampling is not representative of that of the rest of the country. Therefore, a systematic sampling effort of wild boar throughout the country should be included in the national program control. Although in Argentina only the inactivated Bartha vaccine is allowed, recombination risk should not be ignored if attenuated vaccines are incorporated into the National control plan. The two strains, one from the cat and one from the dog sample, are directly related to infected swine. The information about clinical cases and molecular characterization of new strains is important for a better understanding of the dynamics of PRV and to promote preventive measures. Keywords: Argentina, Aujeszky's disease, Pseudorabies. IntroductionPseudorabies virus (PRV), also called Suid herpesvirus type 1, is the causal agent of Aujeszky's disease affecting pigs as primary hosts and reservoirs of the virus. PRV causes several economic losses worldwide in the pig industry (He et al., 2019; Casades-Marti et al., 2020; Klupp, 2021). Eradication efforts in domestic swine were successful in many countries. Several parts of Europe: Austria, Cyprus, the Czech Republic, Denmark, Finland, France, Germany, Hungary, Luxembourg, the Netherlands, Sweden, Switzerland, Slovakia, Norway, and Great Britain; are free of disease in domestic pigs as well as Canada, New Zealand, and the United States. However, in other parts of the world, such as Eastern Europe, Latin America, Asia, and Africa, it is still endemic and is considered a high risk for swine production (Sehl and Teifke, 2020). The emergence of novel PRV strains together with interspecies transmission can generate new strains with a higher potential risk of infection and could represent additional changes to eradication programs (Osterrieder, 2017; Ai et al., 2018; He et al., 2019; Metteinleiter et al., 2019; Yang et al., 2019b; Delva et al., 2020). Pseudorabies is primarily a swine disease, which is considered the natural host, although a diverse range of secondary hosts can become infected and develop the fatal neuronal disease (Casades-Marti et al., 2020). Moreover, the evidence so far suggests that PRV infection may be an occupational risk and could cause disease in humans (Wong et al., 2019). PRV was reported in humans from patients in contact with swine or other domestic animals. Recently, PRV was detected in patients with encephalitis of unknown etiology who had been in contact with swine (Ai et al., 2018; Zhao et al., 2018; Wang et al., 2019; Yang et al., 2019a, 2019b; Sehl and Teifke, 2020). In Argentina, PRV was first detected by Ambrogi et al. (1981). After that, numerous outbreaks were reported in different parts of the country (Davido, 1981; Sager et al., 1984; Echeverría et al., 1991, 1992; Serena et al., 2018). In 2009, the National Veterinary Services (SENASA) implemented an eradication program for PRV (resolution SENASA N° 474/2009). This program includes three stages: 1) Epidemiological studies: prevalence determination and farm classification based on serological studies. Farms were classified as either free of disease, negative, infected, or under a control program. 2) Regionalization: based on the prevalence of positive farms and 3) Control and eradication measures: using vaccination and biosecurity improvement. Genetic farms (those which sell sows, boars, or semen) must be free of the disease determined by negative serological results taken every 120 days. Commercial farms with more than 100 sows must be checked by ELISA every 180 days. Since 2016, SENASA allows domestic swine vaccination with only one type of vaccine: virus-inactivated Bartha k61 gE- strain. Multiple reports describe the detection of PRV in feral pigs and wild boars worldwide. Even in countries where domestic pigs are PRV-free, the virus is almost always present in an endemic form in wild boar (Moreno et al., 2015). In the USA, the successful eradication in domestic swine has been compromised; however, the presence of a reservoir of PRV in the expanding populations of feral swine is a major concern to the pork industry (Corn et al., 2009). The information about seroprevalence in wild boar in Argentina is unknown. Since 2009, SENASA and other institutions have carried out a few studies. One previous report of our group detected 62.5% of seropositive wild boars in 208 sera collected from 2013 to 2015 (Carpinetti et al., 2016). In relation to phylogenetic analysis, there was a clear distinction between the viral strains isolated from the Americas and those isolated from Europe. In previous studies, most of the strains isolated in Argentina were classified as genotype I according to BamHI DNA Restriction fragment length polymorphisms (RFLPs) (Serena et al., 2010). In addition, genomic characterization based on the partial gC gene revealed four different clusters (Serena et al., 2018). At present, in Argentina, PRV cases are sporadically reported. However, providing updated information about clinical cases and molecular characterization of new strains is important for a better understanding of the dynamics of PRV, as well as to promote preventive measures in the region. Materials and MethodsPig distribution and PRV regions according to prevalenceIn the last 10 years, swine production and population have been on a constant increase in Argentina. The Integrated Animal Health Management System (SIGSA) is a SENASA online informatics system that includes information regarding the number of farms, swine population, sanitary events, number of slaughtered animals, etc. Based on the SIGSA database, there are around 5,500,000 pigs, most of them (70%) concentrated in 5 provinces: Buenos Aires (1,338,815 pigs), Córdoba (1,291,783 pigs), Santa Fe (793,434 pigs), Entre Ríos (464,061 pigs) and Chaco (263,473 pigs). The productive structure is represented by the productive units (PUs) which are classified by the number of sows present in each of them. There are around 98,000 PUs in the country, and almost all of them (73%) have 10 or fewer sows each and are considered small producers or subsistence production containing 8.4% of the total pigs in the country. In addition, 24.5% of the total pigs in the country are included in the 0.1% of farms with 10,000 or more pigs. Geographically, Buenos Aires province has the highest percentage of PUs (17.2%) and pigs (24.4%), followed by the provinces of Córdoba (12.6% and 23.6% respectively) and Chaco (13.3% and 4.8% respectively) (Fig. 1a and b).
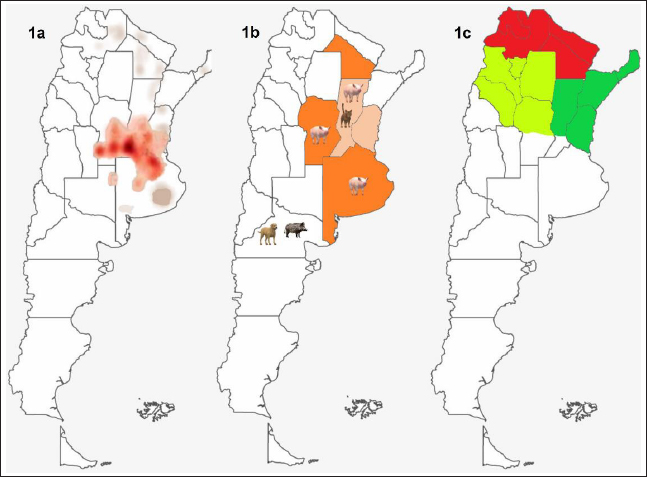
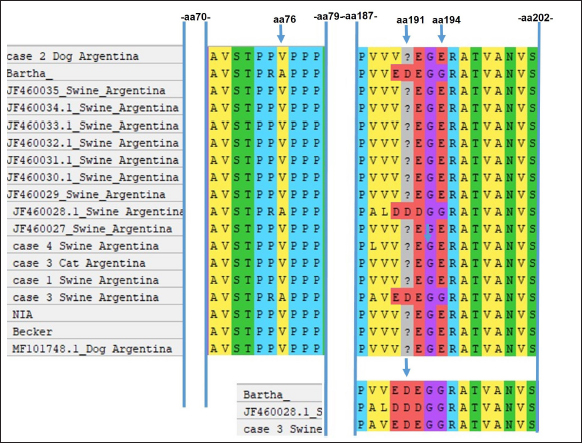
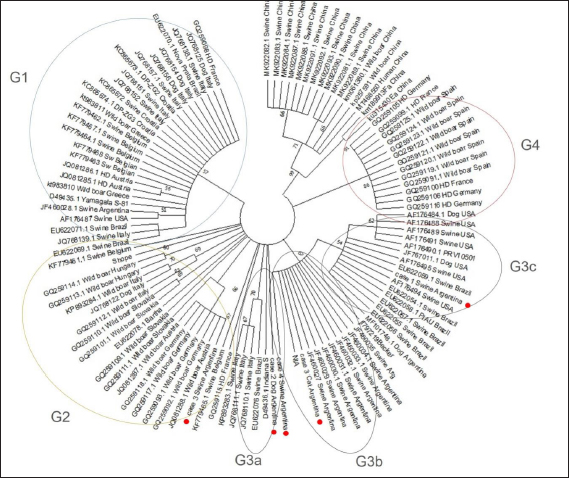
Fig. 1. Swine and farms distribution in Argentina. (a) Swine distribution around the country; (b) the major area of swine production is shown in orange and the area intense orange covers the major number of PUs. Also, the locations of cases detected in this study are indicated by the animal's images. (c) Red and green color indicate the areas of the higher and minor percentages of PRV prevalence, respectively. Evidence of viral circulation in commercial farms is based on antibody detection in the context of the National Control Program. A commercial ELISA (IDEXX PRV/ADV gI or CIVTEST SUIS ADV gE Hipra), which differentiates infected from vaccinated animals, is routinely used. Commercial farms with more than 100 sows must be checked by ELISA every 180 days from samples collected from sows and fattener animals. Thanks to this program, the different regions from the percentage of seroprevalence were detected. For that, a total of 12,919 samples were collected from 1,003 farms, representing 97% of swine stock. Antibodies against PRV in wild boarsFrom 2018 to 2019, 78 wild boars were captured (22 males, 39 females, and 17 not identified) from Bahía de Samborombón natural reserve, located in the Buenos Aires province. Serum samples were analyzed for antibody PRV detection using a virus neutralization test, according to Serena et al. (2018). Recent clinical cases reported in ArgentinaBased on samples submitted to the participating laboratories of this study (Virology of the School of Veterinary Sciences of La Plata, Buenos Aires-INTA Marcos Juárez, Córdoba), from 2013 to 2019, only four positive clinical cases with PRV isolation were detected. All of them were inspected and reported by private practitioners, who collected and submitted the samples to the laboratories. Case 1: Case 1 occurred in 2013 in a 50-sow backyard farm located in the Buenos Aires province. Typical PRV signs were detected in 60-day-old pigs and included neurological signs such as tremors, opisthotonos, and limb stiffness. Samples from the brain and trigeminal ganglia were collected at necropsy and sent to the Virology Laboratory of the School of Veterinary Sciences of La Plata, Buenos Aires. Case 2: Mortality associated with central nervous signs was reported in hunting dogs in the Neuquén Province in 2017. The head of two feral swine and a dog were submitted to the laboratory. Samples from the brain and trigeminal ganglia were analyzed. Case 3: Case 3 was detected in 2019 on a 100-sow farm located in the Santa Fe Province. The clinical case was characterized by the neurological signs in piglets (15 days old) that were detected. A cat from the farm was also found dead. Seven samples of the brain and trigeminal ganglia of piglets and the cat's head were submitted to the laboratory. Case 4: Case 4 was associated with an intensive swine farm. In March 2019, an increase in mortality in fattening pigs was noticed. In the following 2 weeks, mortality also increased in the nursery and weaning pigs. These pigs showed central nervous and respiratory clinical signs (lethargy, opisthotonos, incoordination, anorexia, fever, and dyspnea). The farm owner reported the sudden death of several cats and a dog. The geographical location of each case and PUs by province is presented in Figure 1b. PCR detection and phylogenetic analysisIn each laboratory, samples were individually processed, DNA was extracted using commercial kits (High Pure PCR Template Preparation Kit, Roche Diagnostics, Mannheim, Germany) and PCR was used to detect the gD gene. For sequence analysis, the gC partial gene was amplified as described by Serena et al. (2018). The PCR products were purified according to the manufacturer's protocols using Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI), and sequences on both strands were obtained by the Big Dye Terminator V 3.1 sequencing kit (Applied Biosystems, Germany) with the same primers used for amplification. The sequences were analyzed on an ABI3130XL genetic analyzer (Applied Biosystems, USA), at the Unidad de Genómica, INTA Castelar, Argentina. The sequences obtained were edited using BioEdit software, version 7.2.1. In total, 110 PRV sequences from different geographical locations, including the old Argentine strains, were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/ accession date 06/05/2022). Partial sequences of the gC gene were aligned in the MEGA X program, using the ClustalW algorithm (Kumar et al., 2018). The phylogenetic trees were constructed using the same program as the Maximum Likelihood method and JTT matrix-based model. The nucleotide sequences obtained in this study were submitted to GenBank. ResultsRegions of Argentina according to seroprevalenceThe first two stages of the eradication plan determined that the national prevalence of positive farms was 19.1% and that the disease was more prevalent in small-scale farms. The Northern region of the country including Jujuy, Formosa, Chaco, and Salta provinces, red color in Figure 1c, which is characterized by small-scale or subsistence production farms, and only represents 26% of PUs of Argentina, presented the major proportion of positive farms (45.2%). While a minor prevalence was detected in the two regions colored in green (dark green, 7.4%, and light green, 9.4%, respectively). The region in dark green represents the north of Santa Fe, Entre Rios, Corrientes, and Misiones provinces; and the region in light green includes the north of Cordoba province and Catamarca, La Rioja, Santiago del Estero, and Tucumán provinces. The percentage of farms located in each region is 16.9% and 18.6%, respectively (Fig. 1c). Evidence of PRV circulation in wild boarsFrom the 78 serum samples collected, only 75 were useful for neutralization tests, as 3 of them were contaminated. The seropositivity of wild boars from the samples collected in Bahía de Samborombón was around 41%, most samples which tested positive were female. Phylogenetic and molecular analysisThe new PRV strains identified were confirmed by BLAST analysis, which revealed between 99.74% and 100% similarity to the NIA-3 strain (complete genome KU900059.1). The partial genomic sequences of the PRV gC reported in this paper are available on the NCBI GenBank database (https://www.ncbi.nlm.nih.gov) under accession numbers: OP380625, OP380626, OP380627, OP380628, and OP380629. Sequence analysis revealed maximum nucleotide and amino acid sequence divergence of 3.08% and 12.79%, respectively, compared to the reference strains from GenBank. In addition, we analyzed and compared them at the amino acid level, the four sites involved in adaptive evolution after cross-host transmission among all Argentine strains and the Bartha vaccine strain. Thirteen Argentine strains have P to R change at position 75, V76A, V191D, and E194G. Furthermore, in comparison with NIA-3 and Becker, the Argentine strains were identical to the reference strains. Two Argentine strains, Mer strain (JF460028) and case 3 swine, showed the same amino acids as the Bartha strain (Fig. 2). Phylogenetic analysis of the partial gene encoding the gC protein revealed that the PRV strains have divided into two main clades, clade 1 and clade 2 (Fig. 3). The main difference between clade 1 and clade 2 is the presence of insertion from 59 to 65 amino acids only in Chinese strains belonging to clade 2, that it was also detected in strains isolated from human cases. Clade 1 includes the major number of sequences analyzed and showed four separated groups named: G1, G2, G3, and G4. G1 included the European strains, a few Brazilian strains and a previously reported Argentine strain (JF460028); classified as genotype II by BamHI RFLPs (Serena et al., 2010). G2, G3, and G4 including the RFLP type I strains showed differences in position 24 (S) and 25 (T) amino acids when comparing them with RFLP type II. G2 includes the Bartha vaccine strain, European strains, and one Argentine strain (case 3 swine) that formed a separate group with wild boar strains. In addition, in G3 it is possible to observe three subgroups, named from a to c. Subgroup 3a includes a few strains from Italy and Brazil together with the Indiana S reference strain. The tree topology demonstrates that the new Argentine strains showed no differences from the oldest Argentine PRV strains. Three of the new strains (case 2 dog, case 3 cat, and case 4 swine) are grouped in the same subgroup (3b) with the NIA-3 and Becker strains. One new Argentine strain (case 1 swine) is found more distant from the other Argentine strains and was grouped with the Brazilian strains and the USA strains (subgroup 3c). This subgroup was possibly formed because of an amino acid change (G59P) detected in 3c strains in comparison with 3b strains. Group 4 is formed by some strains from Spain, Germany, and France; all of them have a gap located in the 38–39 amino acid position, which makes them different from other strains which have GG.
Fig. 2. Analysis of amino acid sites involved in the adaptive evolution of PRV. The amino acid position (75, 76, 191, and 194) was analyzed and compared with Bartha, Becker, and NIA-3 reference strains. The changes are shown in the alignment with an arrow. DiscussionThe swine industry has had great growth in Argentina during the last decade. In addition, it couples high-density farming with international trade, generating a higher risk of transmission and potential risk of the global spread of diseases (He et al., 2019). In Argentina, the number of swine has increased significantly, most of the intensive swine farms are located in the major production area of the country (central region). In contrast, about 92% of farms are represented by subsistence, small and medium producers that lack biosecurity and sanitary measures and control. In these kinds of farms, the contact between man, pig, and other domestic and feral animals is very close, which increases the risk of interspecies disease transmission. Nevertheless, these farms represent 20% of the country's pig population (https://www.argentina.gob.ar/sites/default/files/110_1-caracterizacion_porcinos_marzo_2022.pdf). The recent reports of human cases show evidence that people who work with swine, in particular in backyard production, should increase awareness of the risks of not using self-protection (Yang et al., 2019b). The seroprevalence of PRV reported by SENASA is about 19% and it is mainly associated with small-scale farms of 11–50 sows. Besides, the highest seroprevalence was detected in regions with small-scale or subsistence farms (IF-2022-74239096-APN-DPYESA#SENASA). The low seroprevalence detected in commercial farms with more than 100 sows suggests that the national eradication program, based on serological studies, vaccination, and passive vigilance, is in fact working. However, this report informed that the most recent cases of PRV were detected in the central region of our country where pig production is most concentrated. It is important to mention that all the cases came from small-scale farms that lack appropriate biosecurity measures, backyard farms. or wild boars. These findings suggest that the control program should be reformulated and directed to small-scale farms or wild boar reservoirs.
Fig. 3. Phylogenetic analysis of the partial gene encoding the gC protein. Maximum likelihood tree using JTT matrix-based model was constructed by MEGA X software. Groups and subgroups of clade 1 are marked with circular lines. The sequences isolated in this study are indicated with a red dot. In relation to PRV in feral pigs and wild boars, studies from the USA and Europe describe seroprevalence between 0.3% and 66% at the national or regional level depending on the sample size (Müller et al., 2011). In Argentina, neither national nor regional studies have been performed. Since 2009, SENASA evaluated sera samples of a wild boar for PRV detection. However, the number of samples and regions analyzed is low. The study in Bahía de Samborombón revealed a high percentage of detection, but the sampling is not representative of that of the rest of the country. Notwithstanding, it provides important information on PRV circulation in this population. Therefore, a systematic sampling effort of wild boar throughout the country should be included in the national program control. Recombination events have been reported in China in 2011, between wild strains and vaccine strains as well as the re-emergence of PRV in Bartha-vaccinated pig farms (He et al., 2019). Of the 15 strains of PRV reported in Argentina, 13 are closely related to the NIA-3 vaccine strain, and the risk of recombination is possible. Although in Argentina only the inactivated Bartha vaccine is allowed, recombination risk should not be ignored if attenuated vaccines are incorporated into the National control plan. In addition, these potentially recombinant strains may cause virulence enhancement, immune failure, and an increase in cross-host transmission (Skinner et al., 2001). Interspecies transmission has been previously reported in Argentina (Serena et al., 2018). In this report, we detected and characterized two strains, one taken from a cat and one from a dog sample, directly related to infected swine. The genomic characterization of PRV strains originating from swine and other mammals might help better understand the population diversity and facilitate tracing the infection chain back to its origin (Sozzi et al., 2014). The gC gene is a major gene that has been commonly used for the phylogenetic analysis of PRV due to its high variability (Ye et al., 2015). According to other authors, the phylogenetic analysis of partial sequences of the gC gene is a suitable approach for PRV genotyping. In this report, the phylogenetic analysis showed similar patterns as described by others who analyzed the complete gene of gC (He et al., 2019; Zhai et al., 2019). Furthermore, the molecular characterization of new Argentine strains showed no differences from the oldest Argentine PRV strains reported in our previous studies. The Argentine strains formed two different groups inside clade 1 related to NIA-3, Becker, and the USA strains. Moreover, one new Argentinian strain (case 3 swine) is closely related to wild boar strains, suggesting a possible origin from wild pigs. A few amino acid sites were identified with important functions related to the virus infection at cells and four of them are involved in adaptive evolution after cross-host transmission (Karger and Mettenleiter 1993; Flynn and Ryan 1996; He et al., 2019; Zhai et al., 2019). This study involved all amino acid sites of gC described previously that contributed to the grouping of the strains. The case 3 swine in the amino acid positions 75, 76, 191, and 194 showed the same change of amino acid that wild boar strains forming a branch inside of the G2 group. Besides, all these amino acid sites were identical at the Bartha strain. However, case 3 cat maintained the amino acid sequence as older Argentinean strains included in this analysis, suggesting a domestic swine origin. The presence of PRV inside of farm-related to case 3 was demonstrated by the detected virus from animals with clinical signs according to disease. A distinct origin of these two strains is possible because the samples were collected from different animal species. Moreover, when the virus is active in a farm located in a major area of swine production, it is likely to detect different strains. This report provides important information about PRV in Argentina, which may lead to a better understanding of the dynamics of the virus. PRV infection is still present in the country, particularly in regions with small-scale or backyard swine production. This situation will require sanitation and control strategies different from those recommended for intensive high-scale farms. Although PRV outbreaks in swine in Argentina are few, new strains were detected and while the molecular characterization of gC was suitable for carrying out our aim, in the future it will be necessary to include other genes for analysis more exhaustive. This report shows the importance in that continuing with control programs should be emphasized. In particular, programs that aim to eradicate a recent disease is considered zoonotic risk. AcknowledgementsThis work was supported by Proyectos de Incentivos Docentes V260 from Universidad Nacional de La Plata, Argentina; PICT 2019-2756, from Agencia de Promoción Científica y Tecnológica, Argentina and Proyectos PE-I003-INTA and PD-I103- INTA, Instituto Nacional de Tecnología Agropecuaria. Conflict of interestThe authors declare that there is no competing interest regarding the publication of this paper. Author contributionMSS: conceptualization, validation, investigation, formal analysis, writing, review and Editing; JC: methodology and validation, writing, review and editing, supervision, funding acquisition; MF: sampling collection; MW: sampling processing; MD: resources, formal analysis; RB: sampling collection; GEM: investigation and validation; CGA: investigation and validation; AP: review & editing, supervision; BNC: sampling collection (wild boar); MGE: writing, review and editing, Funding acquisition. ReferencesAmbrogi, A., Giraudo, J., Busso, J., Bianco, B., Bagnat, E., Segura de Aramburu, M., Ramos, B. and Ceriatti, S. 1981. Primer diagnóstico de la Enfermedad de Aujeszky en cerdos en la República Argentina. Gac. Vet. 43, 58–64. Ai, J.W., Weng, S.S., Cheng, Q., Cui, P., Li, Y.J., Wu, H.L., Zhu, Y.M., Xu, B. and Zhang, W.H. 2018. Human endophthalmitis caused by Pseudorabies virus infection, China, 2017. Emerg. Infect. Dis. 24, 1087–1090. Carpinetti, B., Castresana, G., Rojas, P., Grant, J., Marcos, A., Monterubbianesi, M., Sanguinetti, H.R., Serena, M.S., Echeverría, M.G., Garciarena, M. and Aleksa, A. 2016. Determinación de anticuerpos contra patógenos virales y bacterianos seleccionados en la población de cerdos silvestres (Sus scrofa) de la Reserva Natural Bahía Samborombón, Argentina. Analecta Vet. 37, 5–11. Casades-Martí, L., González-Barrio, D., Royo-Hernández, L., Díez-Delgado, I. and Ruiz-Fons, F. 2020. Dynamics of Aujeszky's disease virus infection in wild boar in enzootic scenarios. Transbound. Emerg. Dis. 67, 388–405. Corn, J.L., Cumbee, J.C., Barfoot, R. and Erickson, G.A. 2009. Pathogen exposure in feral swine populations geographically associated with high densities of transitional swine premises and commercial swine production. J. Wild. Dis. 45, 713–721. Davido, M. 1981. Enfermedad de Aujeszky en el sur de la Provincia de Córdoba. Gac. Vet. 44, 291–296. Delva, J.L., Nauwynck, H.J., Mettenleiter, T.C. and Favoreel, H.W. 2020. The attenuated Pseudorabies virus vaccine strain Bartha K61: A brief review on the knowledge gathered during 60 years of research. Pathogens (Basel, Switzerland) 9, 897. Echeverría, M.G., Nosetto, E.O., Petruccelli, M., Gimeno, E. and Etcheverrigaray, M. 1991. Ocurrencia y diagnóstico de la Enfermedad de Aujeszky en las zonas de Chañar Ladeado y Saladillo. Vet. Arg. 8, 252–257. Echeverría, M., Nosetto, E., Etcheverrigaray, M., Galosi, C., Fonrouge, R., Pereyra, N., Belak, K. and Gimeno, E. 1992. Pseudorabies (Aujeszky's disease) in Argentina. Rev. Sci. Tech. 11, 819–827. Flynn, S.J. and Ryan, P. 1996. The receptor-binding domain of pseudorabies virus glycoprotein C is composed of multiple discrete units that are functionally redundant. J. Virol. 70, 1355–1364. Karger, A. and Mettenleiter, T.C. 1993. Glycoproteins gIII and gp50 play dominant roles in the biphasic attachment of pseudorabies virus. Virol. 194, 654–664. He, W., Auclert, L.Z., Zhai, X., Wong, G., Zhang, C., Zhu, H., Xing, G., Wang, S., He, W., Li, K., Wang, L., Han, G. Z., Veit, M., Zhou, J. and Su, S. 2019. Interspecies transmission, genetic diversity, and evolutionary dynamics of Pseudorabies Virus. J. Infect. Dis. 219, 1705–1715. Klupp B.G. 2021. Pseudorabies virus infections. Pathogens (Basel, Switzerland) 10(6), 719. Kumar, S., Stecher, G., Li M., Knyaz, C. and Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. Metteinleiter, T.C., Ehlers, B., Muller, T., Yoon, K.J. and Teifke, P. 2019. Herpesviruses. In Diseases of swine, 11th ed. Eds., Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W. and Zhang J. Hobojke, NJ: Wiley & sons, pp: 548–575. Moreno, A., Sozzi, E., Grilli, G., Gibelli, L.R., Gelmetti, D., Lelli, D., Chiari, M., Prati, P., Alborali, G.L., Boniotti, M.B., Lavazza, A. and Cordioli, P. 2015. Detection and molecular analysis of Pseudorabies virus strains isolated from dogs and a wild boar in Italy. Vet. Microbiol. 177, 359–365. Müller, T., Hahn, E.C., Tottewitz, F., Kramer, M., Klupp, B.G., Mettenleiter, T.C. and Freuling, C. 2011. Pseudorabies virus in wild swine: a global perspective. Arch. Virol. 156, 1691–1705. Sager, R., Rossanigo, C., Vázquez, R., Avila, J. and Fondevila, N. 1984. Enfermedad de Aujeszky en cerdos en Villa Mercedes (San Luis) Argentina. Rev. Med. Vet. 65, 86–89. Osterrieder, K. 2017. Herpesvirales. In Fenner´s veterinary virology, 5th ed. Eds., MacLachlan, N.J. and Dubovi, E.J. London, UK: Academic Press, pp: 199–216. Serena, M.S., Metz, G.E., Martín Ocampos, G., Gambaro, S., Mórtola, E. and Echeverría, M.G. 2010. Characterization of suid herpesvirus 1 field isolates from Argentina. Rev. Arg. Microbiol. 42, 179–183. Serena, M.S., Metz, G.E., Lozada, M.I., Aspitia, C.G., Nicolino, E.H., Pidone, C.L., Fossaroli, M., Balsalobre, A., Quiroga, M.A. and Echeverria, M.G. 2018. First isolation and molecular characterization of Suid herpesvirus type 1 from a domestic dog in Argentina. Open Vet. J. 8, 131–139. Sehl, J. and Teifke, J.P. 2020. Comparative pathology of Pseudorabies in different naturally and experimentally infected species—a review. Pathogens (Basel, Switzerland) 9(8), 633. Skinner, G.R., Ahmad, A. and Davies, J.A. 2001. The infrequency of transmission of herpesviruses between humans and animals; postulation of an unrecognised protective host mechanism. Comp. Immunol. Microbiol. Infect. Dis. 24, 255–269. Sozzi, E., Moreno, A., Lelli, D., Cinotti, S., Alborali, G.L., Nigrelli, A., Luppi, A., Bresaola, M., Catella, A. and Cordioli, P. 2014. Genomic characterization of Pseudorabies virus strains isolated in Italy. Transbound. Emer. Dis. 61, 334–340. Yang, X., Guan, H., Li, C., Li, Y., Wang, S., Zhao, X., Zhao, Y. and Liu, Y. 2019a. Characteristics of human encephalitis caused by pseudorabies virus: a case series study. Int. J. Infect. Dis. 87, 92–99. Yang, H., Han, H., Wang, H., Cui, Y., Liu, H. and Ding, S. 2019b. A case of human viral encephalitis caused by pseudorabies virus infection in China. Front. Neurol. 10, 534. Ye, C., Zhang, Q.Z., Tian, Z.J., Zheng, H., Zhao, K., Liu, F., Guo, J.C., Tong, W., Jiang, C.G., Wang, S.J., Shi, M., Chang, X.B., Jiang, Y.F., Peng, J.M., Zhou, Y.J., Tang, Y.D., Sun, M.X., Cai, X.H., An, T.Q. and Tong, G.Z. 2015. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: evidence for the existence of two major genotypes. Virology 483, 32–43. Ye, C., Guo, J.C., Gao, J.C., Wang, T.Y., Zhao, K., Chang, X.B., Wang, Q., Peng, J.M., Tian, Z.J., Cai, X.H., Tong, G.Z. and An, T.Q. 2016. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 491, 56–63. Wang, Y., Nian, H., Li, Z., Wang, W., Wang, X. and Cui, Y. 2019. Human encephalitis complicated with bilateral Acute retinal necrosis associated with Pseudorabies virus infection: a case report. In. J. Infec. Dis. 89, 51–54. Wong, G., Lu, J., Zhang, W. and Gao, G.F. 2019. Pseudorabies virus: a neglected zoonotic pathogen in humans? Emerg. Microbes Infect. 8, 150–154. Zhai, X., Zhao, W., Li, K., Zhang, C., Wang, C., Su, S., Zhou, J., Lei, J., Xing, G., Sun, H., Shi, Z. and Gu, J. 2019. Genome characteristics and evolution of Pseudorabies virus strains in Eastern China from 2017 to 2019. Virol. Sinica. 34, 601–609. Zhao, W.L., Wu, Y.H., Li, H.F., Li, S.Y., Fan, S.Y., Wu, H.L., Li, Y.J., Lü, Y.L., Han, J., Zhang, W.C., Zhao, Y., Li, G.L., Qiao, X.D., Ren, H.T., Zhu, Y.C., Peng, B., Cui, L.Y. and Guan, H.Z. 2018. Clinical experience and next-generation sequencing analysis of encephalitis caused by Pseudorabies virus. Zhonghua Yi Xue Za Zhi, 98, 1152–1157. | ||
| How to Cite this Article |
| Pubmed Style Serena MS, Cappuccio J, Fossaroli M, Williman MM, Dibarbora M, Brizzio R, Metz GE, Aspitia CG, Perez A, Carpinetti BN, Echeverría MG. Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission. Open Vet J. 2023; 13(4): 419-426. doi:10.5455/OVJ.2023.v13.i4.3 Web Style Serena MS, Cappuccio J, Fossaroli M, Williman MM, Dibarbora M, Brizzio R, Metz GE, Aspitia CG, Perez A, Carpinetti BN, Echeverría MG. Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission. https://www.openveterinaryjournal.com/?mno=122379 [Access: April 25, 2024]. doi:10.5455/OVJ.2023.v13.i4.3 AMA (American Medical Association) Style Serena MS, Cappuccio J, Fossaroli M, Williman MM, Dibarbora M, Brizzio R, Metz GE, Aspitia CG, Perez A, Carpinetti BN, Echeverría MG. Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission. Open Vet J. 2023; 13(4): 419-426. doi:10.5455/OVJ.2023.v13.i4.3 Vancouver/ICMJE Style Serena MS, Cappuccio J, Fossaroli M, Williman MM, Dibarbora M, Brizzio R, Metz GE, Aspitia CG, Perez A, Carpinetti BN, Echeverría MG. Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission. Open Vet J. (2023), [cited April 25, 2024]; 13(4): 419-426. doi:10.5455/OVJ.2023.v13.i4.3 Harvard Style Serena, M. S., Cappuccio, . J., Fossaroli, . M., Williman, . M. M., Dibarbora, . M., Brizzio, . R., Metz, . G. E., Aspitia, . C. G., Perez, . A., Carpinetti, . B. N. & Echeverría, . M. G. (2023) Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission. Open Vet J, 13 (4), 419-426. doi:10.5455/OVJ.2023.v13.i4.3 Turabian Style Serena, María Soledad, Javier Cappuccio, Melisa Fossaroli, Macarena Marta Williman, Marina Dibarbora, Renata Brizzio, Germán Ernesto Metz, Carolina Gabriela Aspitia, Alejandro Perez, Bruno Nicolás Carpinetti, and María Gabriela Echeverría. 2023. Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission. Open Veterinary Journal, 13 (4), 419-426. doi:10.5455/OVJ.2023.v13.i4.3 Chicago Style Serena, María Soledad, Javier Cappuccio, Melisa Fossaroli, Macarena Marta Williman, Marina Dibarbora, Renata Brizzio, Germán Ernesto Metz, Carolina Gabriela Aspitia, Alejandro Perez, Bruno Nicolás Carpinetti, and María Gabriela Echeverría. "Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission." Open Veterinary Journal 13 (2023), 419-426. doi:10.5455/OVJ.2023.v13.i4.3 MLA (The Modern Language Association) Style Serena, María Soledad, Javier Cappuccio, Melisa Fossaroli, Macarena Marta Williman, Marina Dibarbora, Renata Brizzio, Germán Ernesto Metz, Carolina Gabriela Aspitia, Alejandro Perez, Bruno Nicolás Carpinetti, and María Gabriela Echeverría. "Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission." Open Veterinary Journal 13.4 (2023), 419-426. Print. doi:10.5455/OVJ.2023.v13.i4.3 APA (American Psychological Association) Style Serena, M. S., Cappuccio, . J., Fossaroli, . M., Williman, . M. M., Dibarbora, . M., Brizzio, . R., Metz, . G. E., Aspitia, . C. G., Perez, . A., Carpinetti, . B. N. & Echeverría, . M. G. (2023) Characterization of new strains of Pseudorabies virus in Argentina: Detection of interspecies transmission. Open Veterinary Journal, 13 (4), 419-426. doi:10.5455/OVJ.2023.v13.i4.3 |